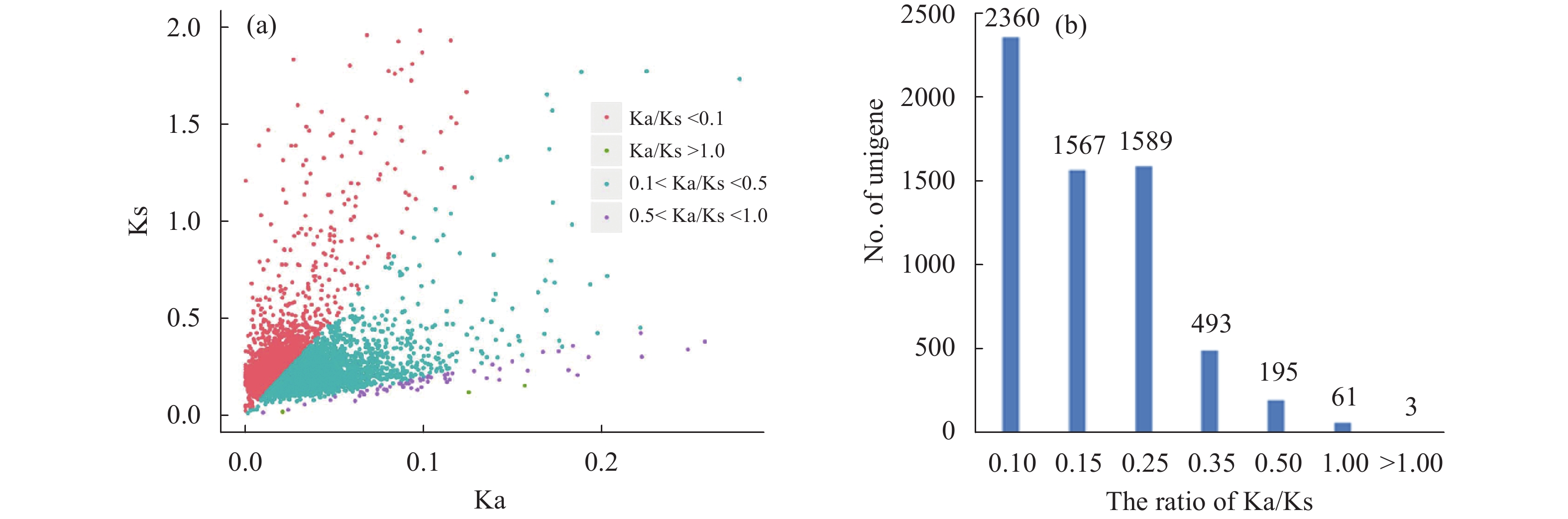
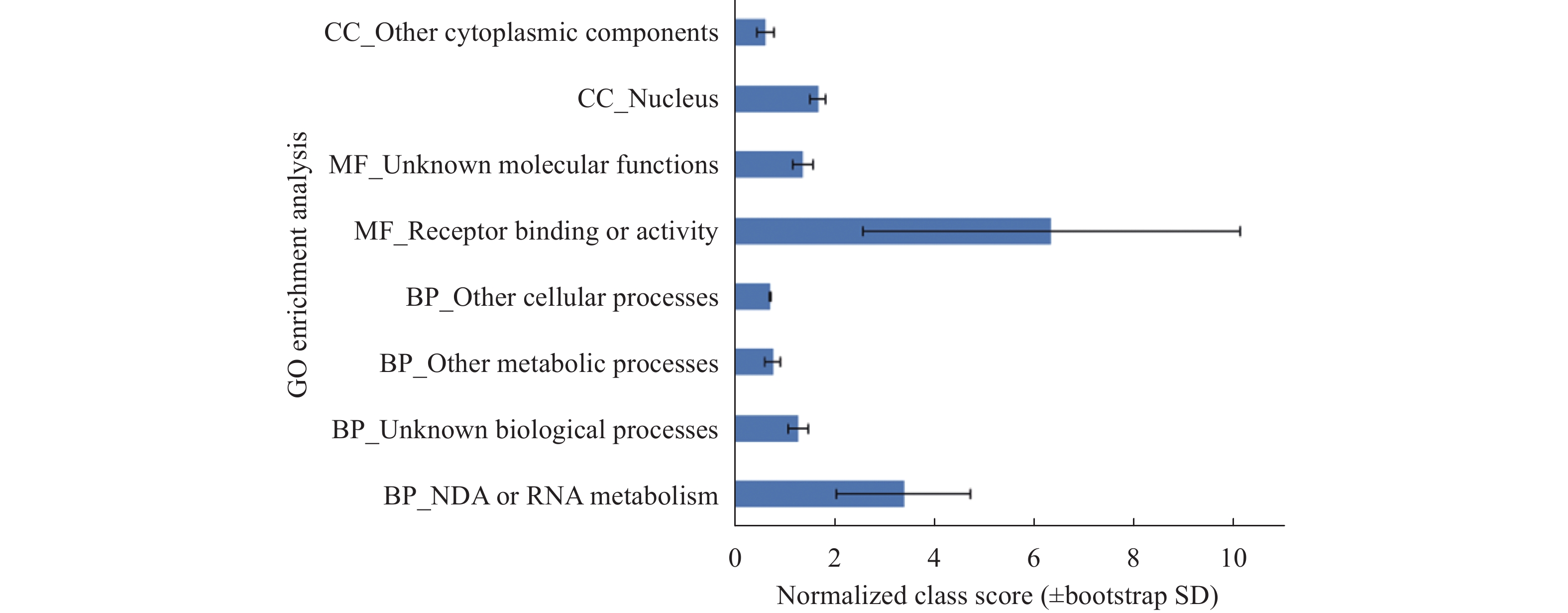
Sciences in Cold and Arid Regions ›› 2018, Vol. 10 ›› Issue (5): 428–435.doi: 10.3724/SP.J.1226.2018.00428
Transcriptomic comparison to identify rapidly evolving genes in Braya humilis
YuMing Wei1,XiaoFei Ma2,PengShan Zhao2,*( )
)
- 1 Animal Husbandry Pasture and Green Agriculture Institute of Gansu Academy of Agricultural Sciences, Lanzhou, Gansu 730070, China
2 Key Laboratory of Stress Physiology and Ecology in Cold and Arid Regions, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu 730000, China
| 1 |
Al-Shehbaz IA, O'Kane Jr SL Arabidopsis gamosepala and A. tuemurnica belong to Neotorularia (Brassicaceae). Novon 1997; 7: 2 93- 94.
doi: 10.2307/3392176 |
| 2 |
Atkin OK, Macherel D The crucial role of plant mitochondria in orchestrating drought tolerance. Annals of Botany 2009; 103: 4 581- 597.
doi: 10.1093/aob/mcn094 |
| 3 |
Bock DG, Andrew RL, Rieseberg LH On the adaptive value of cytoplasmic genomes in plants. Molecular Ecology 2014; 23: 20 4899- 4911.
doi: 10.1111/mec.12920 |
| 4 |
Castresana J Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 2000; 17: 4 540- 552.
doi: 10.1093/oxfordjournals.molbev.a026334 |
| 5 |
Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Research 2015; 43: Database issue D470- D478.
doi: 10.1093/nar/gku1204 |
| 6 |
Chaves MM, Flexas J, Pinheiro C Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany 2009; 103: 4 551- 560.
doi: 10.1093/aob/mcn125 |
| 7 |
Chen LY, Zhao SY, Wang QF, et al. Transcriptome sequencing of three Ranunculus species (Ranunculaceae) reveals candidate genes in adaptation from terrestrial to aquatic habitats . Scientific Reports 2015; 5: 10098.
doi: 10.1038/srep10098 |
| 8 |
Comella P, Pontvianne F, Lahmy S, et al. Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3'ETS in Arabidopsis . Nucleic Acids Research 2008; 36: 4 1163- 1175.
doi: 10.1093/nar/gkm1130 |
| 9 |
Couvreur TLP, Franzke A, Al-Shehbaz IA, et al. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Molecular Biology and Evolution 2010; 27: 1 55- 71.
doi: 10.1093/molbev/msp202 |
| 10 |
Culligan KM, Robertson CE, Foreman J, et al. ATR and ATM play both distinct and additive roles in response to ionizing radiation. The Plant Journal 2006; 48: 6 947- 961.
doi: 10.1111/j.1365-313X.2006.02931.x |
| 11 |
Edgar RC MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 2004; 32: 5 1792- 1797.
doi: 10.1093/nar/gkh340 |
| 12 |
Franzke A, Lysak MA, Al-Shehbaz IA, et al. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Science 2011; 16: 2 108- 116.
doi: 10.1016/j.tplants.2010.11.005 |
| 13 |
Greiner S, Bock R Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays 2013; 35: 4 354- 365.
doi: 10.1002/bies.201200137 |
| 14 |
Hiraguri A, Itoh R, Kondo N, et al. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana . Plant Molecular Biology 2005; 57: 2 173- 188.
doi: 10.1007/s11103-004-6853-5 |
| 15 |
Joseph B, Corwin JA, Li BH, et al. Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2013; 2: e00776.
doi: 10.7554/eLife.00776 |
| 16 |
Kapralov MV, Filatov DA Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evolutionary Biology 2007; 7: 73.
doi: 10.1186/1471-2148-7-73 |
| 17 |
Koenig D, Jiménez-Gómez JM, Kimura S, et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proceedings of the National Academy of Sciences of the United States of America 2013; 110: 28 E2655- E2662.
doi: 10.1073/pnas.1309606110 |
| 18 |
Lasky JR, Des Marais DL, Lowry DB, et al. Natural variation in abiotic stress responsive gene expression and local adaptation to climate in Arabidopsis thaliana . Molecular Biology and Evolution 2014; 31: 9 2283- 2296.
doi: 10.1093/molbev/msu170 |
| 19 |
Lee BH, Henderson DA, Zhu JK The Arabidopsis cold-responsive transcriptome and its regulation by ICE1 . The Plant Cell 2005; 17: 11 3155- 3175.
doi: 10.1105/tpc.105.035568 |
| 20 |
Lee BH, Lee HJ, Xiong LM, et al. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. The Plant Cell 2002; 14: 6 1235- 1251.
doi: 10.1105/tpc.010433 |
| 21 |
Li L, Stoeckert Jr CJ, Roos DS OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Research 2003; 13: 9 2178- 2189.
doi: 10.1101/gr.1224503 |
| 22 |
Liu TM, Tang SW, Zhu SY, et al. Transcriptome comparison reveals the patterns of selection in domesticated and wild ramie (Boehmeria nivea L^ Gaud) . Plant Molecular Biology 2014; 86: 1–2 85- 92.
doi: 10.1007/s11103-014-0214-9 |
| 23 |
Lovell JT, Mullen JL, Lowry DB, et al. Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana . The Plant Cell 2015; 27: 4 969- 983.
doi: 10.1105/tpc.15.00122 |
| 24 |
Mitchell-Olds T, Schmitt J Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis . Nature 2006; 441: 7096 947- 952.
doi: 10.1038/nature04878 |
| 25 |
Moison M, Roux F, Quadrado M, et al. Cytoplasmic phylogeny and evidence of cyto-nuclear co-adaptation in Arabidopsis thaliana . The Plant Journal 2010; 63: 5 728- 738.
doi: 10.1111/j.1365-313X.2010.04275.x |
| 26 |
Niu SH, Li ZX, Yuan HW, et al. Transcriptome characterisation of Pinus tabuliformis and evolution of genes in the Pinus phylogeny . BMC Genomics 2013; 14: 263.
doi: 10.1186/1471-2164-14-263 |
| 27 |
Pastore D, Trono D, Laus MN, et al. Possible plant mitochondria involvement in cell adaptation to drought stress. A case study: durum wheat mitochondria. Journal of Experimental Botany 2007; 58: 2 195- 210.
doi: 10.1093/jxb/erl273 |
| 28 |
Provart NJ, Gil P, Chen WQ, et al. Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiology 2003; 132: 2 893- 906.
doi: 10.1104/pp.103.021261 |
| 29 |
Rizhsky L, Liang HJ, Shuman J, et al. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 2004; 134: 4 1683- 1696.
doi: 10.1104/pp.103.033431 |
| 30 |
Rurek M, Nuc K, Raczyńska KD, et al. Lupin nad9 and nad6 genes and their expression: 5′ termini of the nad9 gene transcripts differentiate lupin species . Gene 2003; 315: 123- 132.
doi: 10.1016/s0378-1119(03)00724-8 |
| 31 |
Schertl P, Braun HP Respiratory electron transfer pathways in plant mitochondria. Frontiers in Plant Science 2014; 5: 163.
doi: 10.3389/fpls.2014.00163 |
| 32 |
Schmid M, Davison TS, Henz SR, et al. A gene expression map of Arabidopsis thaliana development . Nature Genetics 2005; 37: 5 501- 506.
doi: 10.1038/ng1543 |
| 33 |
Sharma S, Villamor JG, Verslues PE Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiology 2011; 157: 1 292- 304.
doi: 10.1104/pp.111.183210 |
| 34 |
Sloan DB Using plants to elucidate the mechanisms of cytonuclear co-evolution. New Phytologist 2015; 205: 3 1040- 1046.
doi: 10.1111/nph.12835 |
| 35 |
Verslues PE, Sharma S Proline metabolism and its implications for plant-environment interaction. The Arabidopsis Book 2010; 8: e0140.
doi: 10.1199/tab.0140 |
| 36 |
Wang LR, Zhao PS, Zhao X, et al. Physiological adaptations to osmotic stress and characterization of a polyethylene glycol-responsive gene in Braya humilis . Acta Societatis Botanicorum Poloniae 2016; 85: 1 3487.
doi: 10.5586/asbp.3487 |
| 37 |
Wang Y, Zhang WZ, Song LF, et al. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiology 2008; 148: 3 1201- 1211.
doi: 10.1104/pp.108.126375 |
| 38 |
Warwick SI, Al-Shehbaz IA, Sauder C, et al. Phylogeny of Braya and Neotorularia (Brassicaceae) based on nuclear ribosomal internal transcribed spacer and chloroplast trnL intron sequences . Canadian Journal of Botany 2004; 82: 3 376- 392.
doi: 10.1139/b04-012 |
| 39 |
Warwick SI, Sauder CA, Al-Shehbaz IA, et al. Phylogenetic relationships in the tribes Anchonieae, Chorisporeae, Euclidieae, and Hesperideae (Brassicaceae) based on nuclear ribosomal its DNA sequences. Annals of the Missouri Botanical Garden 2007; 94: 1 56- 78.
doi: 10.3417/0026-6493(2007)94[56:pritta]2.0.co;2 |
| 40 |
Winter D, Vinegar B, Nahal H, et al. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS One 2007; 2: 8 e718.
doi: 10.1371/journal.pone.0000718 |
| 41 |
Wuest SE, Vijverberg K, Schmidt A, et al. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Current Biology 2010; 20: 6 506- 512.
doi: 10.1016/j.cub.2010.01.051 |
| 42 |
Yang ZH PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 2007; 24: 8 1586- 1591.
doi: 10.1093/molbev/msm088 |
| 43 |
Zhang J, Xie PH, Lascoux M, et al. Rapidly evolving genes and stress adaptation of two desert poplars, Populus euphratica and P^ pruinosa . PLoS One 2013a; 8: 6 e66370.
doi: 10.1371/journal.pone.0066370 |
| 44 |
Zhang L, Yan HF, Wu W, et al. Comparative transcriptome analysis and marker development of two closely related Primrose species (Primula poissonii and Primula wilsonii) . BMC Genomics 2013b; 14: 329.
doi: 10.1186/1471-2164-14-329 |
| 45 |
Zhao PS, Wang LR, Zhao X, et al. A comparative transcriptomic analysis reveals the core genetic components of salt and osmotic stress responses in Braya humilis . PLoS One 2017; 12: 8 e0183778.
doi: 10.1371/journal.pone.0183778 |
| 46 | Zheng D The system of physico-geographical regions of the Qinghai-Xizang (Tibet) Plateau. Science in China (Series D) 1996; 39: 4 410- 417. |
| No related articles found! |