Article Information
- Constantine Uwaremwe, ShiWeng Li, XiMing Chen, Maurice Ngabire, Tawheed Mohammed Elheesin Shareef, Juan Li, MingHui Wu, GuoLi Li . 2017.
- An Arthrobacter strain isolated from desert soils in the region of Shule River (China) can convert cellulose to potential biofuels
- Sciences in Cold and Arid Regions, 9(2): 167-174
- http://dx.doi.org/10.3724/SP.J.1226.2017.00167
Article History
- Received: December 8, 2016
- Accepted: February 22, 2017
2. Key Laboratory of Extreme Environmental Microbial Resources and Engineering, Northwest Institute of Eco-Environment and Resource, Chinese Academy of Sciences, Lanzhou, Gansu 730070, China
Recently, world energy consumption has grown rapidly because of industrialization across the economies (Izhar et al., 2013). There are two main sources of energy used in the world today, renewable and non-renewable. Non-renewable sources such as coal or oil will eventually be exhausted and can seriously pollute the environment and affect our health (Hossain, 2012). Some renewable resources such as ethanol and biogas provide an attractive alternative and may be less polluting (Izhar et al., 2013). Biofuel technology is being upgraded in several countries and research is rapidly increasing, with biofuels eventually providing a significant contribution to solving the world's energy problems (Ottinger, 2007; Gadonneix et al., 2010). In various developed countries, biofuel technology is being explored for use in the transport sector. There are several reasons why biofuel technology could be of a special interest in developing countries as it requires only simple local materials and may be labour intensive (Larson, 2008).
Present-day production of biofuels by agricultural means may itself have some serious environmental consequences. Such concerns include forest destruction to provide land for growing crops such as maize or oil palm, and use of genetically modified crops for increasing yield with its attendant possible genetic consequences for other crops and weed species (Ottinger, 2007).
To overcome such problems, there has been a recent increase in research interest in hydrocarbon producing bacteria. In the biofuel domain, there are numerous bacteria species which have been reported to produce hydrocarbons (Janice et al., 2009). Some microorganisms are able to produce different types of hydrocarbons that contain a carbon number between C 27–C 33 with a double bond in the chain (David et al., 2010a). Aliphatic hydrocarbons produced from cellulose would have the advantage of being suitable for replacing petroleum-based gasoline or diesel in modern engines (Beller et al., 2009).
A microbe's ability to produce hydrocarbon chains is attributed to the presence of special Ole ABCD genes and they have been classified into families on the basis of containing certain enzymes such as thialase, α/β-hydrolase, AMP-dependent ligase/synthase, and short-chain dehydrogenase. Long chain olefinic hydrocarbon producing microorganisms are placed in the families Verucomicrobia,Planctomyces, Chloroflexi, Proteobacteria, and Actinobacter to which Arthrobacter species belong (David et al., 2010b). The Arthrobacter bacteria are gram-positive bacteria, characterized by a high content of DNA G+C, are common inhabitant of the soil environment, leaf surfaces, and several other environments (Zhang et al., 2010; Ganzert et al., 2011; Kallimanis et al., 2011; Scheublin et al., 2012; Saxena et al., 2013; Krishnan et al., 2016). Arthrobacter and other microorganisms are able to generate different potential biofuels from direct decomposition or fermentation using cellulose as their main carbon source (Mazzoli et al., 2012; Du et al., 2015) and this ability might be enhanced by the effect of mutagenesis.
The main objective of this study was to isolate Arthrobacter species having a strong ability to convert cellulose potential biofuel. Furthermore, the mutagenesis method was used to improve the biofuel production of the isolates.
2 Material and methods 2.1 Soil sampling and strain isolationSix soil samples designated SLP1, SLP2, SLP3, SLP4, SLP8, SLP9 were collected from different areas of the Shule River Valley located in Gansu Province, China. After collection, all samples were stored at 4 °C in sterile containers. For each sample, 0.5 g was taken and diluted six-fold in distilled water. Serial dilutions were made and 150 μL samples were spread on plates containing 10 g/L CMC, 15 g/L agar, 1 g/L KNO 3, 0.5 g/L NaCl, 0.5 g/L K 2HPO 4·3H 2O, 0.5 g/L MgSO 4·7H 2O, 0.01 g/L FeSO 4, and pH of 6.8. 1 mL of nalidixic acid was added in order to inhibit the growth of organisms other than Arthrobacter sp. The plates were incubated at 30 °C and after 10 days colonies had formed on most of the plates.
2.2 Assay method of cellulose degradationA qualitative assay was used to evaluate strain ability to degrade cellulose. By using an inoculating needle, a small amount of each colony was taken colony by colony and suspended in 10 μL of distilled water. 1 μL of the suspension for each was used to culture at least 15 colonies on a CMC solid medium plate and soluble starch solid medium containing 15 g/L Agar KNO 3, 0.5 g/L NaCl, 0.5 g/L K 2HPO 4·3H 2O, 0.5 g/L MgSO 4·7H 2O, and 0.01 g/L FeSO 4.
2.3 Biofuel production assay and its chemical property identificationBiofuel production ability was quantitatively analysed by evaluating the quantity in grams of extracted biofuel in 1 g of wet weight for each strain. Strains that show good growth on both CMC-agar medium and soluble starch-agar (SS-Agar) medium were used. For each strain, a small amount was suspended in 1,000 μL of distilled water with 200 μL of the suspension spread on eight plates: 4 LB-Agar plates and 4 SS-Agar plates and incubated at 25 °C. LB-Agar medium was prepared by using 10 g/L NaCl, 10 g/L pryptone, 5 yeast-extract, and 15 agar. After five days of incubation, strains were grown on all plates and collected in sterile containers plate by plate. For each strain, biofuel was extracted using methanol and hexane in a proportion of 1:5. Reagents were added to all strain containers and centrifuged at 8,000 rpm for 15 minutes. The hexane layer was then removed for soxhlet extraction ( Castro et al., 2000; Beller et al., 2009; Redfern et al., 2014). Chemical properties of extracted biofuel were analysed with gas chromatography and mass spectrometry in the laboratory of the Chinese Academy of Science, Gansu Province, China.
2.4 Strain identification, 16S rRNA gene sequencing and phylogenetic analysisIsolated strains were identified by means of morphological examination using an optical microscopy to identify which strain had Arthrobacter morphology. For further identification, the chromosomal DNA of the isolates was extracted using E.Z.N.A Bacterial DNA Kit and the DNA fragment of rRNA gene was amplified. The PCR products were sequenced in the Huada Gene Technology Ltd (Beijing). The obtained sequences were aligned using Fasta and phylogenetic analysis was carried out using MEGA5.5.
2.5 MutagenesisFor further enhancing biofuel productivity of the isolates, the mutagenesis using N-methyl-N-nitrosoguanidine (NTG) was conducted. The strain was suspended in phosphate buffer saline (PBS) pH 7.2 and 5 μL of NTG was added, then incubated at 30 °C for 30 minutes after it was washed with PBS. Serial dilutions of the strain were prepared and cultured on CMC-Agar plates for ten days. All mutant strains were later analyzed for their biofuel production abilities by growing them on LB-Agar plates.
2.6 Optimization of biofuel production conditions of mutant strainsTo determine biofuel production conditions, the four mutant strains that produced more biofuel were grown in different media and incubated at 30 °C. For each mutant strain, biofuel was extracted medium by medium in two different durations of culture.
2.7 Statistical analysisData were analysed using Microsoft Excel. The t-test was done by making a comparison of the mean values for individual variables for each strain between the two conditions at 99% significance level. For all the six strains, data were analysed by one-way analysis of the variance (one-way ANOVA). By using Duncan's multiple range tests at an α-risk of 5%, we estimated the difference of every considered variable.
3 Results and discussion 3.1 Isolation and identification of the strain with strong ability to degrade celluloseIn this study, 57 strains were isolated according to their colony diameter on the CMC-Agar medium. Among them, 32 strains were identified morphologically as belonging to the genus Arthrobacter. In a total of 32 strains, growth ability on SS-Agar plate was 100% but only 18.57% on CMC-Agar. Qualitative assay of cellulose degradation revealed that 26 strains show good growth on SS-Agar medium and poor growth on CMC-Agar medium, whereas, six strains show good growth on both media (Figure 1, Table 1).
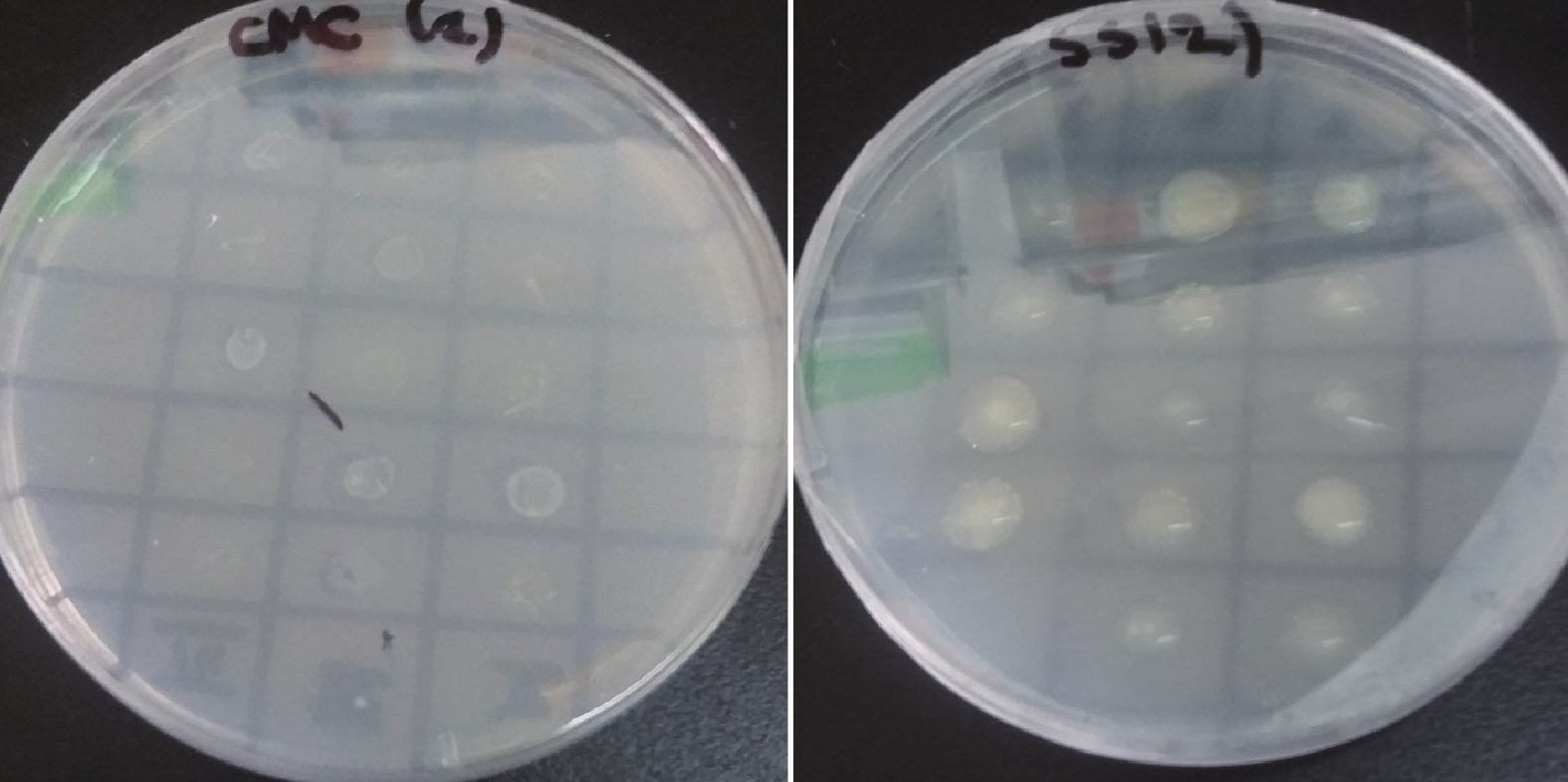
|
| Figure 1 Growth ability of strains on CMC-Agar and soluble starch SS-Agar medium. On SS-Agar plate all strains show good growth, whereas on CMC-Agar plate few were well developed |
| Strain | Morphological characterization | Colony diameter (mm) |
| SLP4-1 | Rod-coccus, V-shape | 3.5 |
| SLP9 | Rod-coccus, V-shape | 4.0 |
| SLP8 | Rod-coccus, V-shape | 3.0 |
| SLP2 | Rod-coccus, V-shape | 3.5 |
| SLP1 | Rod-coccus, V-shape | 3.0 |
| SLP4-2 | Rod-coccus, V-shape | 4.0 |
Qualitative assay of cellulose degradation show that only six strains with a colony diameter between 3~4 mm show good growth on CMC-Agar medium and these strains were used for the biofuel production assay (Table 1). In recent studies, numerous bacteria have been reported as cellulose degrading bacteria isolated using the transparent zone selection (Avellaneda et al., 2014). For example, in a cellulose degradation study of Bacillus brevis it was shown that the formation of a transparent zone on culture medium around the colony indicated cellulose degradation (Fatema et al., 2015). Other cellulose degrading bacteria isolated from mangrove forest show a similar clear halo around the colony on CMC medium (Behera et al., 2014).
3.2 Biofuel production and analysis of biofuel chemical compositionWe further evaluated the biofuel production by six strains that show good growth on CMC-Agar and SS-Agar plates. Results show that only one strain (SLP1) had the ability to produce a higher content of biofuel in the fermentation broth (Figure 2). Results from gas chromatographic and mass spectroscopic analysis of the extracted biofuel are presented in Table 2. The chemical structures of the hydrocarbons with the five highest peaks are presented in Table 3.
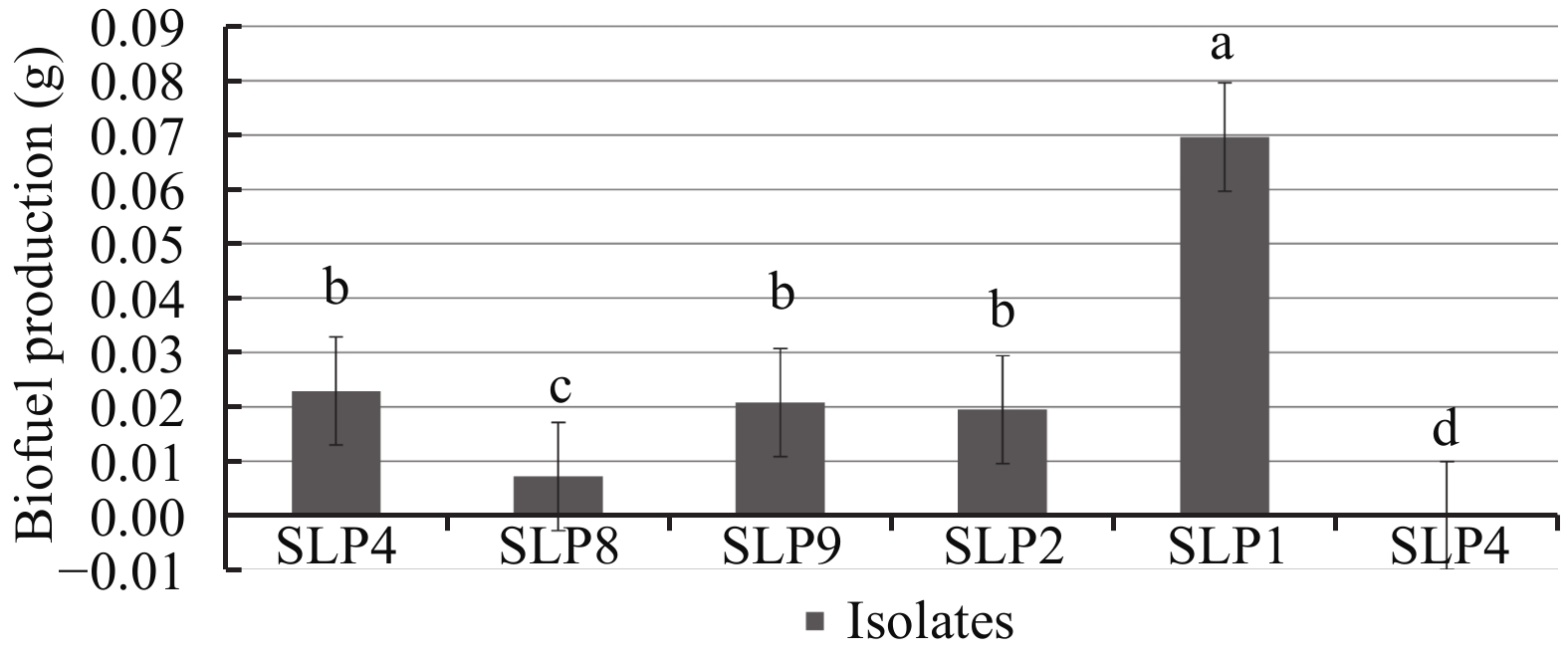
|
| Figure 2 The ability of biofuel production by the six isolates. Post-test analyses were done by Duncan's multiple range test (p<0.01) using Microsoft-Excel; different letters on bars indicate the significance atp<0.05 |
| No. | Detected hydrocarbons | Signal strength | Retention time (min) |
| 1 | Dodecane, Heneicosane, Hexadecane | 0.98 | 9.296 |
| 2 | 2-Bromododecane, Tridecane, 1-iodo-Hexadecane | 3.68 | 9.730 |
| 3 | 2,4-bis (1,1-dimethylethyl) Phenol, 2,4-bis (1,1-dimethylethyl); Phenol, 2,4-bis (1,1-dimethylethyl) | 2.80 | 10.029 |
| 4 | 2-Bromo dodecane, Heneicosane, Tridecane, 1-iodo- | 1.30 | 10.267 |
| 5 | 2-Bromotetradecane, Heneicosane, Tetratriacontane | 1.04 | 11.373 |
| 6 | Heptadecane, 3-methyl-Octadecane, Pentadecane, 2,6,10-trimethyl- | 1.03 | 11.509 |
| 7 | Heptadecane, 9-octyl-Octadecane, Octadecane | 1.46 | 11.944 |
| 8 | Heptadecane | 3.04 | 12.086 |
| 9 | Heneicosane, 2-Bromotetradecane, Heptadecane | 4.66 | 12.229 |
| 10 | Heneicosane, Heptadecane | 1.46 | 12.690 |
| 11 | 1-Octadecene, 1-Nonadecene, Pentadecyl -trifluoroacetate | 7.85 | 13.111 |
| 12 | Octadecane | 1.59 | 13.166 |
| 13 | Heneicosane, Pentacosane | 3.49 | 13.695 |
| 14 | Eicosane, 10-methyl-Heneicosane, Octadecane | 1.32 | 14.089 |
| 15 | Heptadecane, 3-methyl-Triacontane, Tetracosane | 1.12 | 14.130 |
| 16 | Pentadecane, 8-hexyl-Eicosane, 10-methyl-Octadecane | 1.88 | 14.197 |
| 17 | Octadecane, 1-iodo-Eicosane | 0.90 | 14.387 |
| 18 | Heneicosane, Octacosane, Dodecane, 2,6,11-trimethyl- | 5.76 | 14.476 |
| 19 | Octadecane, Heneicosane, Pentacosane | 1.90 | 14.890 |
| 20 | 5-Eicosene, 3-Eicosene, (E)- 1-Docosene | 1.80 | 15.127 |
| 21 | Eicosane | 0.96 | 15.182 |
| 22 | Heptadecane, 3-methyl-Heneicosane, Tetratriacontane | 1.08 | 15.786 |
| 23 | Tetratriacontane, Hentriacontane | 1.22 | 16.057 |
| 24 | Heneicosane | 2.20 | 16.125 |
| 25 | Tetratetracontane, Tetratriacontane, Octacosan | 1.11 | 16.248 |
| 26 | Octadecane, 1-iodo-Hexacosane, Heptadecane, 2-methyl- | 2.20 | 16.397 |
| 27 | Heptacosane, Heneicosane, Hexadecane, 2-methyl- | 4.52 | 16.506 |
| 28 | Tetratriacontane, Heptacosane, Hentriacontane | 2.09 | 16.791 |
| 29 | Octadecane, 1-iodo-Hentriacontane, Octacosane | 1.31 | 16.872 |
| 30 | Behenic alcohol, 1-Heneicosanol, Pentadecyl pentafluoropropionate | 10.86 | 17.001 |
| 31 | Docosane, Hexadecane, 2,6,10,14-tetramethyl-Docosane | 2.55 | 17.035 |
| 32 | Heneicosane, Octadecane, Octacosane | 1.73 | 18.311 |
| 33 | Hexadecane, 2-methyl-Tetratriacontane, Octadecane, 1-iodo- | 2.35 | 18.359 |
| 34 | 4-([(Furan-2-ylmethyl)-amino]-methyl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl ester, Benzeneethanamine, N[(pentafluorophenyl)methylene]-4-[(trimethylsilyl)oxy]- Benzo[h]isoquinoline | 1.85 | 18.427 |
| 35 | 9-Octadecenamide, (Z)-9-Octadecenamide, (Z)-9-Octadecenamide | 2.94 | 18.569 |
| 36 | Heptadecane, Octadecane, 3-methyl-Octadecane, 5,14-dibutyl- | 1.15 | 19.391 |
| 37 | Nonadecane, Tricosane, Heptadecane, 2-methyl- | 1.13 | 19.520 |
| 38 | Hentriacontane, Octadecane, 1-iodo-Octacosane | 2.05 | 20.002 |
| 39 | Octacosane, Hentriacontane | 0.89 | 20.056 |
| 40 | Octadecane, Hentriacontane, Heptadecane, 3-methyl- | 1.24 | 21.848 |
| 41 | 1-Docosanol, methyl ether Eicosane, 9-cyclohexyl-Hexatriacontyl pentafluoropropionate | 5.52 | 22.330 |
Five hydrocarbons were detected at highest peaks eluted in time distance of 12.229~22.330 min from C 16–C 22 belonging chemically to alkane, n-alkene, and alcohol families, respectively (Table 3). Hydrocarbon syntheses from numerous bacteria have been studied by other authors. Ladygina et al. (2006) reported intracellular production of hydrocarbons contains C 18–C 27 and C 25–C 35 production by Clostridium, a gram-positive bacterium which basically grows by a fermentative process. In yeast, hydrocarbon synthesis depended considerably on the growth condition (Ladygina et al., 2006). In another study, Janice et al. (2009) reported the long-chain monoalkenes containing cis-3,25-dimethyl-13-heptacosene were produced by an Arthrobacter species, indicating that Arthrobacter was able to produce hydrocarbons as biofuels.
| No. | Retention time (min) | Signal strength | Molecule structure |
| 9 | 12.229 | 4.66 |

|
| 11 | 13.111 | 7.85 |
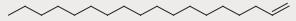
|
| 18 | 14.476 | 5.76 |
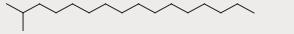
|
| 30 | 17.001 | 10.86 |
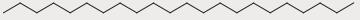
|
| 41 | 22.330 | 5.52 |
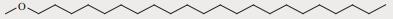
|
Morphological characteristics of the isolate SLP1 analysed by optical microscopic observation (Table 1) shows that this isolate had the general features of the genus Athrobacter. The identification of the isolate SLP1 was further identified by its 16S rRNA gene sequence. A phylogenetic tree was constructed through its 16S rRNA gene as presented in Figure 3. The results indicate that the isolate SLP1 was phylogenetically related to other members of the genus Arthrobacter, and thus is referred to as Athrobacter nitroguajacolicus.

|
| Figure 3 Neighbour-joining tree based on the 16S rRNA gene sequence of the strain SLP1 and the sequences of representative strains from the Gene Bank. The bar represents 0.0002 substitutions per site respectively |
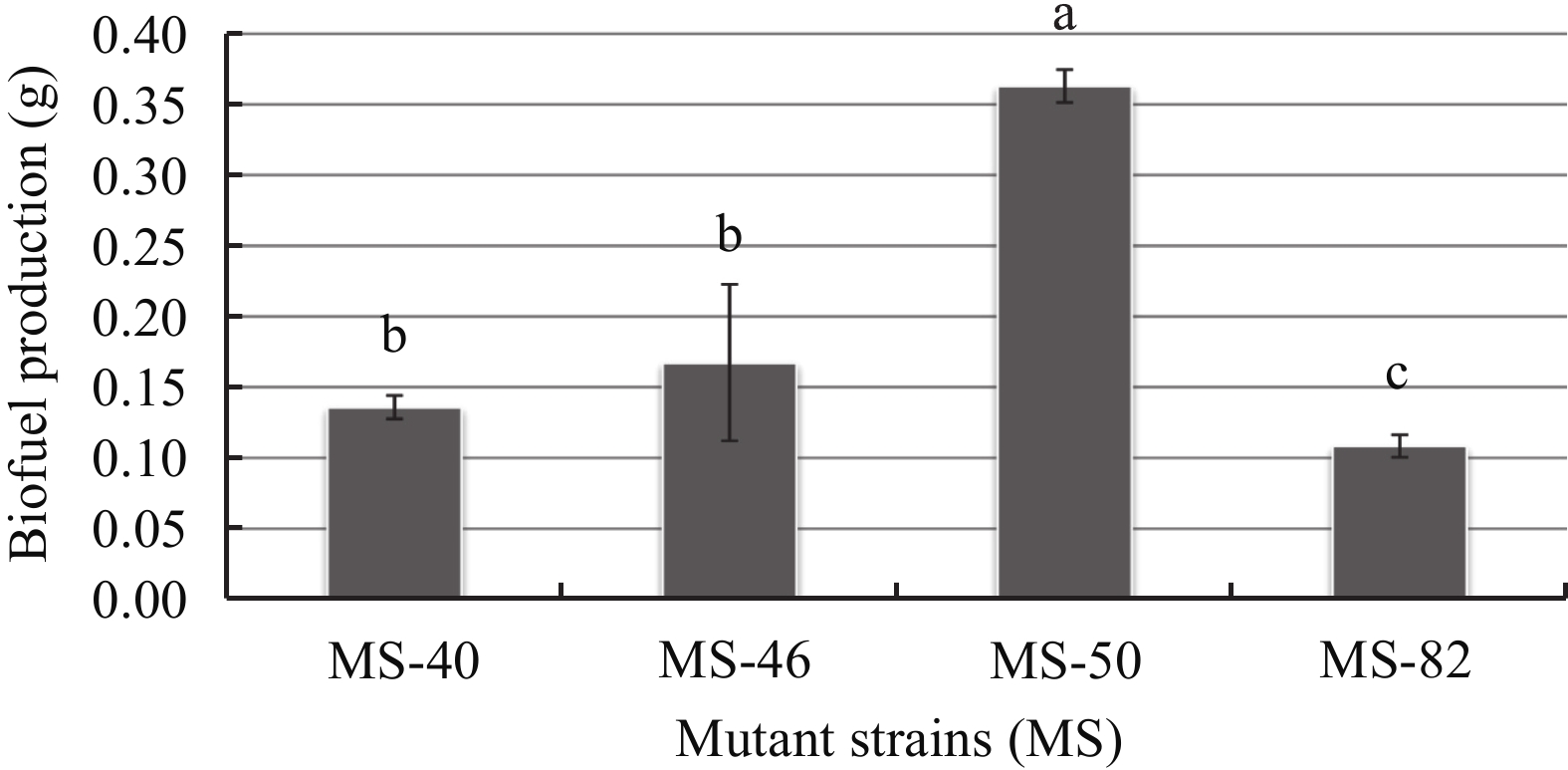
After NTG mutagenesis, we obtained 94 mutant strains that had higher ability for biofuel producing relative to the original strain. Among them, four strains show greater biofuel production ability than the original strain before mutation. Quantitative analyses of the extracted biofuel shows that the amount of the biofuel in the fermentation broth produced by the MS-40, MS-46, MS-50, and MS-82 mutant strains were increased by 95.67%, 141.21%, 422.62% and 56.19%, respectively (Figure 4). The result indicates that after mutagenesis, the biofuel production had enhanced considerably. Numerous studies have reported the enhancement of biofuel production using different methods in microbes. For example, genetic engineering was used to enhance biofuel production in microalgae (Fatemeh et al., 2015). In Thermophile sp. ethanol production was enhanced by classical methods including selection of clones and nonspecific mutagenesis (Scull et al., 2015).
|
| Figure 4 Biofuel productions of four mutant strains in the LB-Agar medium |
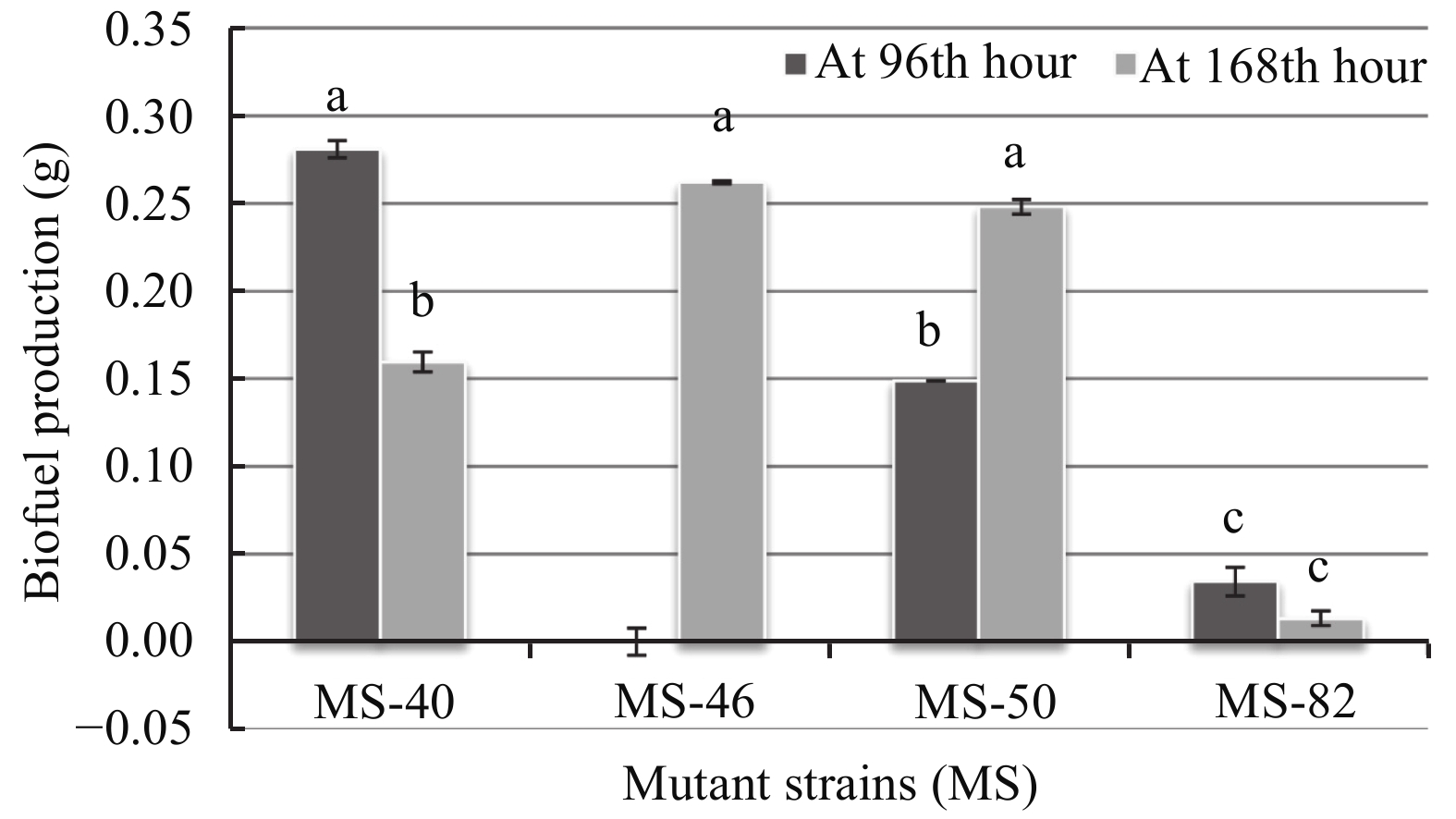
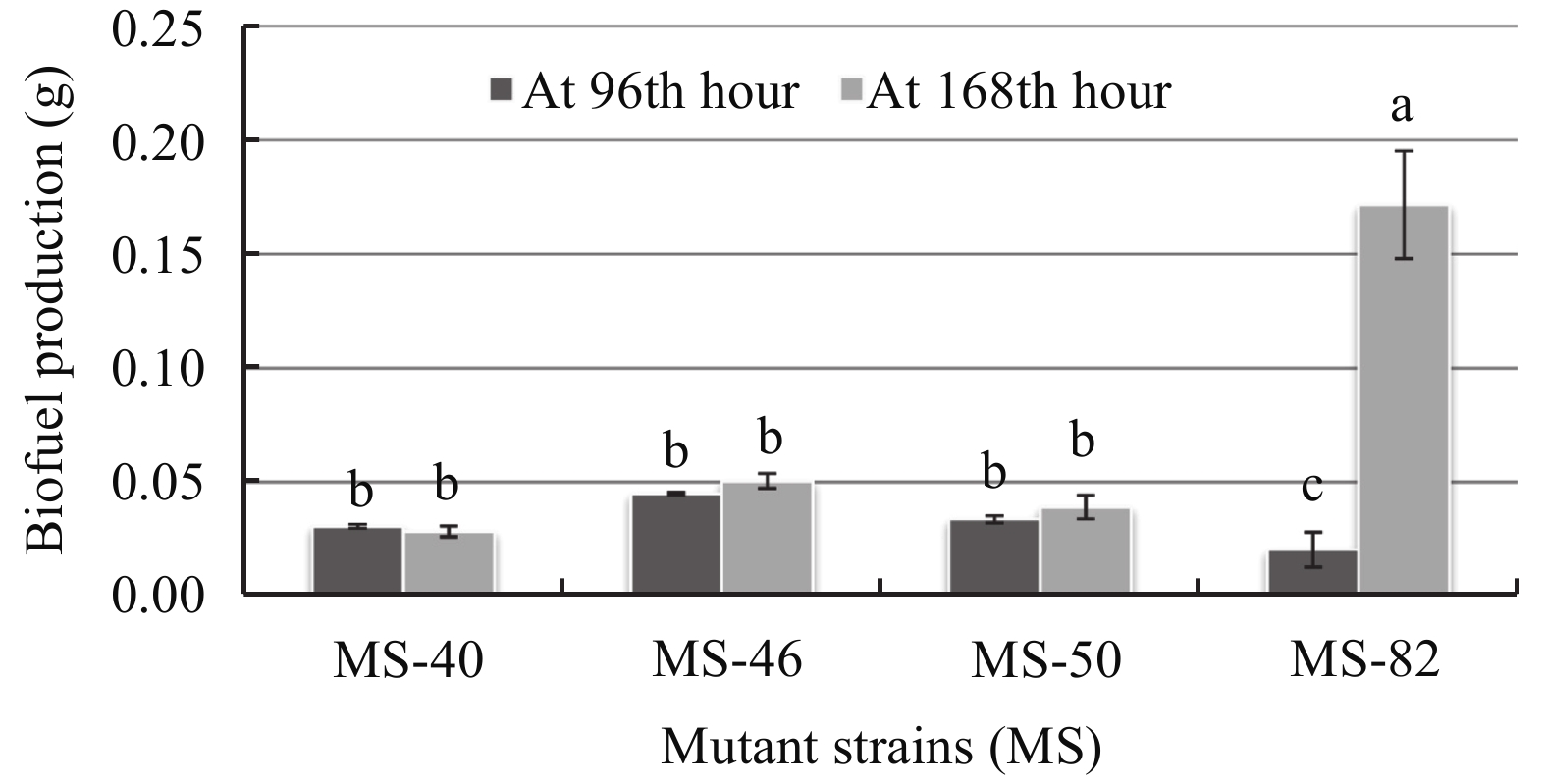
LB-Agar medium and SS-Agar medium were chosen to optimize biofuel production of the four mutant strains at different times of culture. Results show that strain biofuel production ability was medium-dependant and time-dependant for all mutants (Figures 5 and 6). When grown on SS-Agar medium, mutant strain MS-40 produced more biofuel at the 96th hour whereas it was moderate at the 168th hour. Mutant MS-46 shows a large biofuel production at the 168th hour whereas there was no biofuel production at the 96th hour. Mutant MS-50 show significant biofuel production at the 96th hour and a further significant increase at the 168th hour. Mutant strain-82 shows no significant production at both 96th hour and 168th hour. For MS-40, MS-46, and MS-50 there was little significant difference between time and biofuel production, whereas a significant difference was observed with a 9.32 fold increase between the 96th and 168th hour for MS-82.
|
| Figure 5 Biofuel production of the mutant strains on SS-Agar medium |
|
| Figure 6 Biofuel production of the mutant strains on LB-Agar medium |
In this study, we isolated Arthrobacter strains that could grow in CMC-Agar medium which exhibited the ability to produce biofuel from cellulose; the most productive strain was identified asArthrobacter nitroguajacolicus strain SLP1. The identified hydrocarbons in the fermentation broth of this strain were alkane, alkene, and alcohols with carbon numbers ranging from 16 to 22 (C 16–C 22). Further mutagenesis result indicates that biofuel productivity of the strain was greatly enhanced. From this study, it can be concluded that Arthrobacter nitroguajacolicus isolated from soils in the region of Shule River in Gansu Province, China, could efficiently produce hydrocarbons which might be used as energy sources in the future.
Acknowledgments:This study was financially supported by the National Natural Science Foundation of China (31400437, 31560121), the international cooperation program of Gansu (1504WKCA097), the application transformation foundation of CAS (HHS-CGZH-16-02) and UK BBSRC China Partnering Grant (BB/J020419/1).
| Avellaneda LM, Pulido CPG, Rojas ET, 2014. Assessment of cellulolytic microorganisms in soils of Nevados Park, Colombia. Brazilian Journal of Microbiology, 45: 1211–1220. DOI: 10.1590/S1517-83822014000400011 |
| Behera BC, Parida S, Dutta SK, et al, 2014. Isolation and identification of cellulose degrading bacteria from mangrove soil of Mahanadi River Delta and their cellulase production ability. American Journal of Microbiological Research, 2(1): 41–46. DOI: 10.12691/ajmr-2-1-6 |
| Beller HR, Goh EB, Keasling JD, 2009. Genes involved in long-chain alkene biosynthesis inMicrococcus luteus . Applied and Environmental Microbiology, 76: 1212–1223. |
| Castro L, Ayuso G, 2000. Soxhlet Extraction. Environmental Application. Academic Press, pp. 2701–2709. |
| David JS, Jennifer L, Jack E, et al, 2010a. Structure, function, and insights into the biosynthesis of a head-to-head hydrocarbon in Shewanella oneidensis strain MR-1 . Applied Environmental Microbiology, 76: 3842–3849. DOI: 10.1128/AEM.00433-10 |
| David JS, Jennifer LS, Jack ER, et al, 2010b. Widespread head-to-head hydrocarbon biosynthesis in bacteria and the role of OleA. Applied Environmental Microbiology, 76: 3850–3862. DOI: 10.1128/AEM.00436-10 |
| Du R, Yan J, Li S, et al, 2015. Cellulosic ethanol production by natural bacterial consortia is enhanced by Pseudoxanthomonas taiwanensis . Biotechnology for Biofuels, 8(10): 1–10. |
| Fatema K, Manchur MA, 2015. Isolation, identification and cellulase production by Bacillus brevis from the Acacia Forest soil . International Journal of Research in Agriculture and Forestry, 2: 14–22. |
| Fatemeh N, Jamshid R, 2015. Genetic engineering of microalgae for enhanced biodiesel production suitable fuel replacement of fossil fuel as a novel energy source. American Journal of Life Sciences, 3: 32–41. |
| Ganzert L, Bajerski F, Mangelsdorf K, et al, 2011. Arthrobacter livingstonensis sp. nov. and Arthrobacter cryotolerans sp. nov., salt-tolerant and psychrotolerant species from Antarctic soil . International Journal of Systematic and Evolutionary Microbiology, 61: 979–984. DOI: 10.1099/ijs.0.021022-0 |
| Gadonneix P, de Castro FB, de Medeiros NF, et al., 2010. Biofuels: Policies, Standards and Technologies. United Kingdom. |
| Hossain KA, 2012. Global energy consumption pattern and GDP. International Journal of Renewable Energy Technology Research, 1: 23–29. |
| Izhar Q, Fahimuddin S, 2013. Analysis of global energy consumption patterns. Journal of Environmental Research and Development, 7: 1–13. DOI: 10.1016/j.envdev.2013.05.013 |
| Janice A, Richman JE, Wackett LP, 2009. C29 olefinic hydrocarbons biosynthesized by Arthrobacter species . Applied and Environmental Microbiology, 75: 1774–1777. DOI: 10.1128/AEM.02547-08 |
| Kallimanis K, LaButti AM, Lapidus A, et al, 2011. Complete genome sequence of Arthrobacter phenanthrenivorans type strain (Sphe3) . Standards in Genomic Sciences, 4: 123–130. DOI: 10.4056/sigs.1393494 |
| Krishnan R, Menon RR, Tanaka N, et al, 2016. Arthrobacter pokkalii sp. nov., a novel plant associated Actinobacterium with plant beneficial properties, isolated from saline tolerant pokkali rice, Kerala, India . PLoS ONE, 11(3): e0150322. DOI: 10.1371/journal.pone.0150322 |
| Ladygina N, Dedyukhina EG, Vainshtein MB, 2006. A review on microbial synthesis of hydrocarbons. Process Biochemistry, 41: 1001–1014. DOI: 10.1016/j.procbio.2005.12.007 |
| Larson ED, Trade UNC, 2008. Biofuel production technologies: status, prospects and implications for trade and development. United Nations Conference on Trade and Development, New York and Geneva. |
| Mazzoli R, 2012. Development of microorganisms for cellulose-biofuel consolidated bioprocessings: metabolic engineers' trick. Computational and Structural Biotechnology, 3(4): 2–10. |
| Ottinger RT, 2007. Biofuels - Potential, Problems and Solutions. Biofuels Conference Rio de Janeiro, pp. 16–19. |
| Redfern J, Kinninmonth M, Burdas D, et al, 2014. Using soxhlet ethanol extraction to produce and test plant material (essential oils) for their antimicrobial properties. Journal of Microbiology & Biology Education, 15: 45–46. |
| Scull SM, Orlygsson J, 2015. Recent advances in second generation ethanol production by thermophilic bacteria. Energies, 8: 1–30. |
| Saxena N, Singh K, 2013. Study of genetic relatedness among arthrobacter strains by phylogenetic analyses of RAPD-PCR fingerprint patterns. International Journal of Advanced Biotechnology and Research, 4: 29–33. |
| Scheublin TR, Leveau JHJ, 2012. Isolation of Arthrobacter species from the phyllosphere and demonstration of their epiphytic fitnes . Microbiology Open, 2: 205–213. |
| Zhang DC, Schumann P, Liu HC, et al, 2010. Arthrobacter alpinus sp. nov., a psychrophilic bacterium isolated from alpine soil . International Journal of Systematic and Evolutionary Microbiology, 60: 2149–2153. DOI: 10.1099/ijs.0.017178-0 |
 2017, 9
2017, 9


