Article Information
- XiaoHui Liang, YuXia Wu. 2017.
- Identification of Kalidium species (Chenopodiaceae) by DNA barcoding
- Sciences in Cold and Arid Regions, 9(1): 89-96
- http://dx.doi.org/10.3724/SP.J.1226.2017.00089
Article History
- Received: July 12, 2016
- Accepted: October 19, 2016
Kalidium Moq. (Chenopodiaceae), are identified as Euhalophytes and divided into five species, Kalidium caspicum (L.), Kalidium gracile Fenzl, Kalidium cuspidatum (Ung.-Sternb.) Grub. var. cuspidatum, Kalidium foliatum (PALL.), Kalidium schrenkianum Bunge. ex Ung. -Sternb and one variant species, Kalidium cuspidatum (Ung.-Sternb.) Grub. var. sinicum A. J. Li, mainly distributed in Southeast Europe and Northwest Asia as shrubs (Kong, 1979; Krever et al., 1998). Previous studies have demonstrated that Kalidium species play an important role in maintaining the balance of grassland ecosystems and preventing soil erosion (Zhao et al., 2002). In comparative studies from three other Euhalophytes species (Suaeda sala, Atriplex centralasiatiea and Nitraria sibirica), Kalidium have proven to possess strong tolerance to saline-alkali soil and drought, as a dominant species in desert areas (Zhao et al., 2002; Zhou et al., 2009). In China, Kalidium is a succulent salt plant mainly used as forage grass for camels, horses and sheep, excellent for grazing herds in the winter. Traditional taxonomic methods for identifying Kalidium relied on morphological and phenotypic characters, which have limits in differentiating species (Kong, 1979). Therefore, developing a common DNA barcoding for species identification of Kalidium is required.
As an increasingly prevalent molecular technique to remedy the limitation of taxonomic research relying solely on morphological features, DNA barcoding has been used to facilitate accurate species-level identification using specific DNA regions (Kress and Erickson, 2007). A potential DNA barcode, CO1 (cytochrome c oxidase subunit 1), was successfully identified as a standard mitochondrial region to discriminate species in animal groups (Hogg and Hebert, 2004; Barrett and Hebert, 2005; Fu et al., 2011). Although previous studies have proposed many potential DNA barcodes, including a nuclear DNA locus (ITS) and several chloroplast DNA regions (matK, rbcL, trnH-psbA, atpF-atpH, et al.) has been widely applied in plant ecology and evolutionary studies (Kress and Erickson, 2007; Newmaster and Ragupathy, 2009; Moniz and Kaczmarska, 2010), universal DNA barcodes for plant species identification have not been very informative (Newmaster et al., 2006; Ford et al., 2009; Dong et al., 2015).
In this study, four candidate DNA loci were selected, one nuclear gene (ITS) and two previously used chloroplast genes (matK and rbcL) and one recently developed candidate chloroplast gene (ycf1b) (Dong et al., 2015), to screen their suitability as DNA barcodes for Kalidium. The major purpose of this study is to address the following two questions: (1) determine ideal DNA markers for species-level identification of Kalidium, (2) verify identification of the variant species of K.cuspidatum var. sinicum is consistent with that of traditional taxonomy relying only on morphology.
2 Material and methods 2.1 Plant samplesLeaf tissue of five species and one variant species (Figure 1) were collected within the full range of Kalidium in China. For each species, 20 to 30 individuals were sampled from each population; and all the samples were silica gel-dried. Three to four individuals were sampled from three to five populations for each species in order to avoid individual bias for this study. A total of 82 samples, representing geographic location and collection information are available in Table 1. We selected Salsola laricifolia Turcz. ex Litv and Chenopodium album L. (Chenopodiaceae) as outgroups.

|
| Figure 1 Morphological characteristics of different Kalidium species. (a) K. schrenkianum, (b) K. cuspidatum var. cuspidatum, (c) K. foliatum, (d)K. gracile, (e) K. caspicum, (f) K.cuspidatum var. sinicum |
| Taxon | Locality | Lon. | Lat. | Alt.(m) |
| K. foliatum(n=15) | Alashanzuoqi, NMG | 39°54′58″E | 105°42′18″N | 1, 019 |
| Minqin, GS | 38°18′32″E | 103°16′32″N | 1, 485 | |
| Balikun, XJ | 43°46′21″E | 91°43′18″N | 1, 630 | |
| Jinghe, XJ | 44°38′34″E | 83°15′10″N | 221.5 | |
| Luntai, XJ | 41°59′34″E | 85°16′07″N | 966 | |
| K.capsicum(n=12) | Manasi, XJ | 44°12′57″E | 86°39′19″N | 514 |
| Mulei, XJ | 43°50′40″E | 90°37′28″N | 854 | |
| Balikun, XJ | 43°46′21″E | 91°43′18″N | 1, 630 | |
| Midong, XJ | 44°07′14″E | 87°42′52″N | 499 | |
| K. gracile (n=15) | Balikun, XJ | 43°38′06″E | 91°57′28″N | 1, 966 |
| Wulatehouqi, NMG | 38°52′11″E | 106°45′23″N | 1, 028 | |
| Dulan, QH | 39°09′19″E | 98°10′14″N | 3, 393 | |
| Pingluo, NX | 40°50′43″E | 103°36′33″N | 1, 095 | |
| Gaolan, GS | 36°26′57″E | 103°59′03″N | 1, 762 | |
| K. schrenkianum (n=10) | Kuche, XJ | 41°55′32″E | 82°51′15″N | 1, 448 |
| Kuche, XJ | 42°06′47″E | 83°08′48″N | 1, 586 | |
| Baicheng, XJ | 41°35′26″E | 81°20′08″N | 1, 487 | |
| K.cuspidatum var. cuspidatum(n=15) | Wulan, QH | 36°42′30″E | 99°02′45″N | 3, 087 |
| Jingtai, GS | 37°20′39″E | 104°05′05″N | 1, 572 | |
| Dingbian, SX | 37°40′11″E | 107°30′56″N | 1, 297 | |
| Pingluo, NX | 38°52′11″E | 106°45′23″N | 1, 095 | |
| Wulateqianqi, NMG | 38°52′22″E | 108°41′54″N | 1, 053 | |
| K. cuspidatum var. sinicum(n=15) | Dulan, QH | 36°01′31″E | 97°38′51″N | 3, 043 |
| Gaolan, GS | 36°27′48″E | 103°55′58″N | 1, 806 | |
| Minqin, GS | 38°18′32″E | 103°16′32″N | 1, 485 | |
| Geermu, QH | 35°51′05″E | 94°31′15″N | 3, 568 | |
| Zhangye, GS | 39°07′44″E | 100°33′04″N | 1, 660 | |
| Abbreviations: n, number of individuals analyzed; Lon., longitude; Lat., latitude; Alt., altitude; GS, Gansu; QH, Qinghai; SX, Shaanxi; XJ, Xinjiang Uygur Autonomous Region; NX, Ningxia Hui Autonomous Region; NMG, Neimenggu Autonomous Region. | ||||
DNA was extracted from leaves by the slightly modified SDS method (Jia et al., 2010). PCR amplification of four candidate barcodes was carried out on the CyclerTM Thermal Cycler (Bio-Rad, USA). Detailed information on the amplification conditions of the tested four regions is available in Table 2. Direct sequencing PCR products were carried out by two directions. Five individuals of the ITSlocus contained segregating indels that prevented direct sequencing, PCR fragments were purified and sub-cloned into the pMD19-T vector (Takara, China) and three to five clones were then directly sequenced.
| Region | Primers | Primer sequence (5ˊ-3ˊ) | Thermocycling conditions | References |
| ITS | ITS4 ITS1 |
TCCTCCGCTTATTGATATGC AGAAGTCGTAACAAGGTTTCCGTAGG |
94 °C, 5 min; [35 cycles: 94 °C, 45 s; 57 °C, 45 s; 72 °C, 80 s]; 72 °C, 10 min |
White et al., 1990 |
| ycf1b | F R |
TCTCGACGAAAATCAGATTGTTGTGAAT ATACATGTCAAAGTGATGGAAAA |
94 °C, 5 min; [35 cycles: 94 °C, 45 s; 55 °C, 45 s; 72 °C, 70 s]; 72 °C, 10 min |
|
| matK | Xf 5r |
TAATTTACGATCAATTCATTC GTTCTAGCACAAGAAAGTCG |
94 °C, 5 min; [35 cycles:94 °C, 45 s; 47 °C, 45 s; 72°C, 90 s]; 72 °C, 10 min |
|
| rbcL | af 724R |
ATGTCACCACAAACAGAGACTAAAGC TCGCATGTACCTGCAGTAGC |
94 °C, 5 min; [35cycles:94 °C, 45 s; 57 °C, 45 s; 72 °C, 70 s]; 72 °C, 10 min |
Kress et al., 2005 |
Sequence chromatograms were edited and aligned using Aligner v.5.1.0 (Codon Code Corporation, Dedham, MA), with all posterior probabilities > 0.8 and all polymorphic and heterozygous sites manually confirmed. We calculated K2P (Kimura 2-parameter) distances using MEGA 5 software to evaluate the intra-specific and inter-specific differences (Kumar et al., 2008). The distribution graphs of intra-specific and inter-specific K2P genetic distances of each DNA locus were made and compared to value barcode gaps using Origin 8.5 (Meier et al., 2006). The differentiation power of the four DNA barcodes was estimated using tree-based methods. Bootstrap support values were performed through 1, 000 random replicates.
3 Results 3.1 PCR amplification, sequencing and alignmentAll four candidate loci exhibited high PCR success and sequencing success (100%) (Table 3). The 336 new sequences of the four DNA regions were obtained representing five species and one variant of Kalidium, which also included two outgroup taxa, S.laricifolia and C. album, respectively (Table 3). The results showed that the tested primers possessed prominent universality (Table 3). GeneBank accession numbers of all the sequences are listed in Table 4.
| DNA region | ITS | ycf1b | matK | rbcL |
| Universal ability to primer | 100 | 100 | 100 | 100 |
| Sequencing success rate (%) | 100 | 100 | 100 | 100 |
| Length of sequence (bp) | 681~686 | 881/899 | 902 | 696 |
| Aligned sequence length (bp) | 693 | 899 | 902 | 696 |
| No. indels | 11 | 1 | 0 | 0 |
| Indel length (bp) | 1~4 | 18 | 0 | 0 |
| No. of variable sites | 119 | 35 | 7 | 7 |
| No. sampled species (individuals) |
82 | 82 | 82 | 82 |
| Interspecific distance mean (range) |
0.0437 (0.014~0.066) | 0.0061 (0.000~0.013) | 0.0030 (0.000~0.006) | 0.0045 (0.000~0.009) |
| Intraspecific distance mean (range) |
0 | 0.0019 (0.000~0.009) | 0.0004 (0.000~0.004) | 0.0004 (0~0.006) |
| Ability to distinguish (%) | 100.0 | 16.7 | 33.3 | 16.7 |
| Species | GenBank accession number | |||
| ITS | matK | rbcL | ycf1b | |
| K. caspicum | KX133017-KX133028 | KX133101-KX133112 | KX133185-KX133196 | KX133269-KX133280 |
| K. cuspidatum var. cuspidatum | KX133029-KX133043 | KX133113-KX133127 | KX133197-KX133211 | KX133281-KX133295 |
| K. cuspidatum var. sinicum | KX133044-KX133058 | KX133128-KX133142 | KX133212-KX133226 | KX133296-KX133310 |
| K. foliatum | KX133059-KX133073 | KX133143-KX133157 | KX133227-KX133241 | KX133311-KX133325 |
| K. gracile | KX133074-KX133088 | KX133158-KX133172 | KX133242-KX133256 | KX133326-KX133340 |
| K. schrenkianum | KX133089-KX133098 | KX133173-KX133182 | KX133257-KX133266 | KX133341-KX133350 |
| C. album | KX133016 | KX133100 | KX133184 | KX133268 |
| S. laricifolia | KX133099 | KX133183 | KX133267 | KX133351 |
For four DNA loci, aligned sequence lengths possessed a relatively great range 681 bp for ITS to 902 bp for matK (Table 3). The DNA barcode containing the most variable sites was ITS (119), followed by ycf1b(35), rbcL (7) and matK(7). The distribution graphs of the intra-specific and inter-specific distance for the four DNA regions displayed the highest mean inter-specific divergences of ITS region (0.0437), and matK had the lowest mean inter-specific divergences (0.003) (Table 3).
3.2 DNA barcoding gapGraphing the distribution of K2P distances is to evaluate the barcoding divergence between intraspecific and interspecific genetic distances for all DNA barcodes tested (Figure 2). We found a large barcoding gap only in the ITS region and the other three DNA sequences tested proved to have no such barcoding gap (Figure 2).
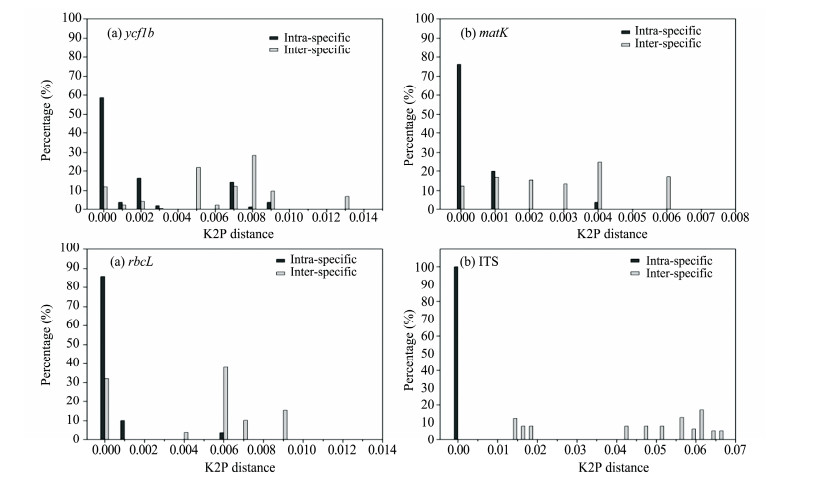
|
| Figure 2 Relative distribution photographs of inter-specific and intra-specific distances for the four DNA barcodes of Kalidium. The x-axes stands for K2P distances arranged in intervals and the y-axes stands for the percentage of occurrences |
Neighbor-Joining (NJ) trees were constructed with the single-locus or combined DNA loci to evaluate the discrimination power for the four DNA barcodes (Figures 3, 4). As the single locus analyses, the species discrimination power of the ITS locus was highest, with a success rate of 100%, followed by matK (33.3%), ycf1b (16.7%) and rbcL (16.7%) (Table 3). The tree-based method for the ycf1b locus identified the same species as that of the rbcL locus (Figure 4). For possible combination of the other three DNA loci excluding ITS region analyses, the results showed that the possible combinations of matK with the other two loci had the same performance as using matK alone (Figure 4). Due to the relatively low success of species identification for single locus or random combination of the tested three barcodes excluding ITS region, the confident tree obtained only from ITS (Figure 3).

|
| Figure 3 Neighbor-joining tree based on the ITS gene sequences with the Kimura 2-parameter distance model. Bootstrap values are available above the relevant branches and species are shown in the right column |
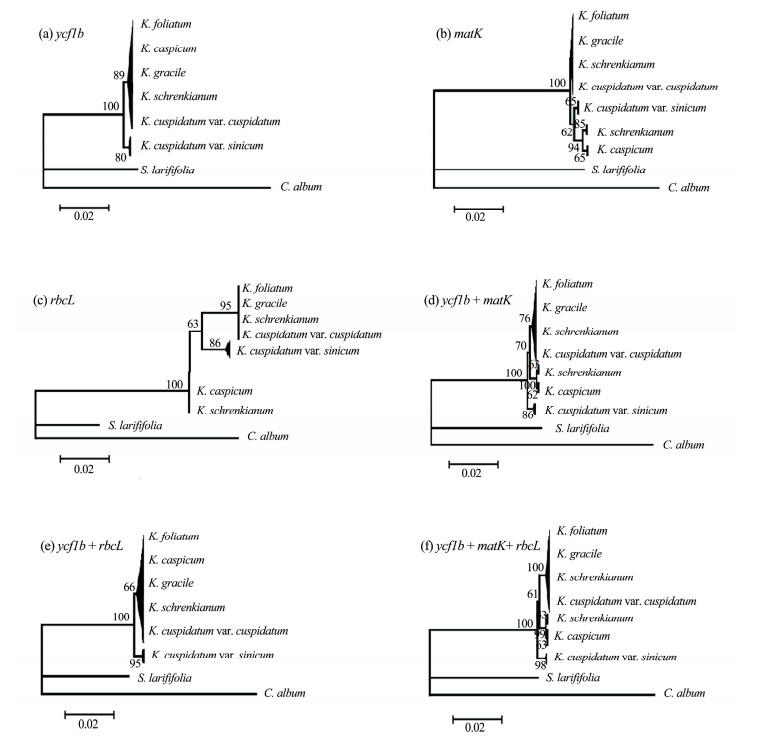
|
| Figure 4 Neighbor-joining phylogram based on the three DNA regions (matK, rbcL, ycf1b) with the Kimura 2-parameter distance model. Bootstrap values are available above or below the relevant branches and the values lower than 50% were covered |
NJ tree analysis of ITS sequences revealed that the 82 samples used in this study were significantly split into six clades (Figure 3). Single or any combination of all four DNA barcodes tested identified K.cuspidatum var. sinicum as a distinctive clade (Figures 3, 4). The analysis of all NJ trees identified K.cuspidatum var. sinicum as a single species with high bootstrap values.
4 Discussion and conclusionsThe success rate of PCR amplification and sequencing has long been treated as a significant index to estimate DNA barcodes. In this study, all four DNA barcodes tested were universal with PCR amplification and sequencing success. Through sequences analysis, the ITS region exhibited high resolution, as identification power has been showed in Alnus (Ren et al., 2010) and Euphorbiaceae (Pang et al., 2010). For species level identification, ITS showed great potential for being an ideal DNA barcode for Kalidium due to its high inter-specific divergence and discrimination rate (100%) in the four DNA loci tested. rbcL, as one of the core DNA barcodes for plants, has performed well for mosses, ferns and angiosperms (Hollingsworth et al., 2009; Liu et al., 2011; Zheng et al., 2015). Also, previous studies have indicated that the rbcLregion showed relatively low inter-specific K2P distances on determining closely related species (Hasebe et al., 1995; Newmaster et al., 2008; Gong et al., 2015). Our results showed that rbcL had relatively low inter-specific distance and the lowest species differentiation rate, and as such is not suitable as a DNA locus for discrimination in Kalidium. Recently, Dong et al. (2015) demonstrated that ycf1b is a plastid genome region with relatively high variable sites and proposed ycf1b as a core DNA barcode for species identification of land plants. Our results showed that ycf1b had the same discrimination rate (16.7%) as rbcL and were unfit for potential DNA barcode in Kalidium. The matK region has proven to possess a relatively significant inter-specific K2P distances and good generality in some land plants (Kelly et al., 2010; Gong et al., 2015). Lahaye et al. (2008) identified matK gene showed prominent universality for identification of flower plants species. Asahina et al. (2010) demonstrated that matK rather than rbcL was better suited in identifying medicinal Dendrobium species due to a high-level resolution. The matK locus showed relatively high level of discrimination rate in Kalidium species compared with the other two candidates (rbcL and ycf1b, the matK locus was proposed as a candidate DNA barcode rather than an ideal barcode owning to its lower species level identification than the ITS region in Kalidium.
Numerous studies have demonstrated the limitation of species delimitation relying solely on morphological features. For example, Protoparmelia was more diverse than what was expected from traditional taxonomy, consisting of several previous unknown depicted species, and cryptic species-lineages (Singh et al., 2015). According to morphological and biogeographic information, Wood (2006) treated Dendrobium officinale and D. tosaenseas a common species, but by molecular technique identification results suggested that they should be identified as two different species (Asahina et al., 2010). Compared to K. cuspidatum var. cuspidatum, K. cuspidatum var. sinicum was identified as a variant species based solely on different morphological traits (Kong, 1979). According to our observations on morphological characteristics of these two species in the field, there were significant differences on growth of the shoots, the length and diameter of the infructescences. Also, the phylogenetic analyses of all four DNA barcodes tested in this study showed that K. cuspidatum var. sinicum clustered into a single clade with strong bootstrap values greatly separated from the other five species. Obviously, species identification of Kalidium relying solely on morphology has its limitations. Thus, we suggested that K. cuspidatum var. sinicum should be identified as a single species in Kalidium, rather than as a variant.
In conclusion, results showed that the ITS region was an ideal DNA barcode in Kadilium, and the variant, K. cuspidatum var. sinicum, should be treated as a single species.
Acknowledgments:We are grateful to KuiBing Meng, DeCheng Liu and FengZhu Zhang for sample collections in the field. This work was supported by the Program for New Century Excellent Talents in the Ministry of Education in China (NCET-09-0446), and lzujbky-2012-k22 to YuXia Wu.
| Asahina H, Shinozaki J, Masuda K, et al, 2010. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. Journal of Natural Medicine, 64(2): 133–138. DOI: 10.1007/s11418-009-0379-8 |
| Barrett RDH, Hebert PDN, 2005. Identifying spiders through DNA barcodes. Canadian Journal of Zoology, 83(3): 481–491. DOI: 10.1139/z05-024 |
| Dong WP, Chao X, Chang HL, et al, 2015. ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports, 5: 1–5. DOI: 10.1038/srep08348 |
| Ford CS, Ayres KL, Toomey N, et al, 2009. Selection of candidate coding DNA barcoding regions for use on land plants. Botanical Journal of the Linnean Society, 159(1): 1–11. DOI: 10.1111/j.1095-8339.2008.00938.x |
| Fu YM, Jiang WM, Fu CX, 2011. Identification of species within Tetrastigma (Miq. ) Planch. (Vitaceae) based on DNA barcoding techniques. Journal of Systematics and Evolution, 49(3): 237–245. DOI: 10.1111/j.1759-6831.2011.00126.x |
| Gong W, Liu Y, Chen J, et al, 2015. DNA barcodes identify Chinese medicinal plants and detect geographical patterns of Si-nosenecio (Asteraceae). Journal of Systematics and Evolution, 54(1): 83–91. DOI: 10.1111/jse.12166 |
| Hasebe M, Wolfe PG, Pryer KM, 1995. Fern phylogeny based on rbcL nucleotide sequences. American Fern Journal, 85(4): 134–181. DOI: 10.2307/1547807 |
| Hogg ID, Hebert PDN, 2004. Biological identification of spring-tails (Hexapoda:Collembola) from the Canadian Arctic, using mitochondrial DNA barcodes. Canadian Journal of Zoology, 82(5): 749–754. DOI: 10.1139/z04-041 |
| Hollingsworth ML, Clark AA, Forrest LL, et al, 2009. Selecting barcoding loci for plants:evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Molecular Ecology Resources, 9(2): 439–457. DOI: 10.1111/j.1755-0998.2008.02439.x |
| Jia J, Cai L, Shi C, 2010. Study on extraction method of nucleic acid in Kalidium foliatum (Pall. ). Chinese Agricultural Science Bulletin, 26(4): 49–52. |
| Kelly LJ, Ameka GK, Chase MW, 2010. DNA barcoding of African Podostemaceae (river-weeds):a test of proposed barcode re-gions. Taxon, 59(1): 251–260. |
| Kong XW, 1979. Flora of China. Beijing:Science Press, pp. 14, 16. |
| Kress WJ, Erickson DL, 2007. A two-locus global DNA barcode for land plants:the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE, 2(6): e508. DOI: 10.1371/journal.pone.0000508 |
| Krever V, Pereladova O, Williams M, et al., 1998. Biodiversity Conservation in Central Asia:An Analysis of Biodiversity and Current Threats and Initial Investment Portfolios. World Wild Fund for Nature (WWF), Almaty. |
| Kumar S, Nei M, Dudley J, et al, 2008. MEGA:a biologist-centric software for evolutionary analysis of DNA and protein se-quences. Briefings in Bioinformatics, 9(4): 299–306. DOI: 10.1093/bib/bbn017 |
| Lahaye R, van der Bank M, Bogarin D, et al, 2008. DNA barcoding the floras of biodiversity hotspots. Proceedings of the National Academy of Sciences of the United States of America, 105(8): 2923–2928. DOI: 10.1073/pnas.0709936105 |
| Liu J, Moller M, Gao LM, et al, 2011. DNA barcoding for the discrimination of Eurasian yews (Taxus L. , Taxaceae), and the discovery of cryptic species. Molecular Ecology Resources, 11(1): 89–100. DOI: 10.1111/j.1755-0998.2010.02907.x |
| Meier R, Shiyang K, Vaidya G, et al, 2006. DNA barcoding and taxonomy in Diptera:a tail of high intraspecific variability and low identification success. Systematic Biology, 55(5): 715–728. DOI: 10.1080/10635150600969864 |
| Moniz MB, Kaczmarska I, 2010. Barcoding of diatoms:nuclear encoded ITS revisited. Protist, 161(1): 7–34. DOI: 10.1016/j.protis.2009.07.001 |
| Newmaster SG, Fazekas AJ, Ragupathy S, 2006. DNA barcoding in land plants:evaluation of rbcL in a multigene tiered approach. Canadian Journal of Botany, 84(3): 335–341. DOI: 10.1139/b06-047 |
| Newmaster SG, Fazekas AJ, Steeves RAD, et al, 2008. Testing candidate plant barcode regions in the Myristicaceae. Molecular Ecology Resources, 8(3): 480–490. DOI: 10.1111/j.1471-8286.2007.02002.x |
| Newmaster SG, Ragupathy S, 2009. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Molecular Ecology Resources, 9(Supp. 1): 172–180. DOI: 10.1111/j.1755-0998.2009.02642.x |
| Pang XH, Song JY, Zhu YJ, et al, 2010. Using DNA barcoding to identify species within Euphorbiaceae. Planta Medica, 76(15): 1784–1786. DOI: 10.1055/s-0030-1249806 |
| Ren BQ, Xiang XG, Chen ZD, 2010. Species identification of Alnus (Betulaceae) using nrDNA and cpDNA genetic markers. Mo-lecular Ecology Resources, 10(4): 594–605. DOI: 10.1111/j.1755-0998.2009.02815.x |
| Singh G, Dal Grande F, Divakar PK, et al, 2015. Coalescent-based species delimitation approach uncovers high cryptic diversity in the cosmopolitan lichen-forming fungal genus Protoparmelia (Lecanorales, Ascomycota). PLoS ONE, 10(5): e0124625. DOI: 10.1371/journal.pone.0124625 |
| Wood HP, 2006. The Dendrobiums. Ruggell:Gantner Verlag, pp. 847. |
| Zhao KF, Fan H, Jiang XY, et al, 2002. Improvement and utilization of saline soil by planting halophytes. Chinese Journal of Applied & Environmental Biology, 8(1): 31–35. |
| Zheng SH, Ren WG, Wang ZH, et al, 2015. Use of chloroplast DNA barcodes to identify Osmunda japonica and its adulterants. Plant Systematic and Evolution, 301(7): 1843–1850. DOI: 10.1007/s00606-015-1197-y |
| Zhou ZY, Yan SY, Qin Y, et al, 2009. The character of shrub diversity in arid desert regions in Alashan. Journal of Arid Land Resources and Environment, 23(9): 146–150. DOI: 10.13448/j.cnki.jalre.2009.09.025 |
 2017, 9
2017, 9


