Article Information
- ChaoJu Qian, MengHe Gu, HengXia Yin, Yong Shi, ChengLiang Yin, Xia Yan, XiaoFei Ma . 2017.
- Adaptive evolution of rbcL in Reaumuria soongarica (Tamaricaceae)
- Sciences in Cold and Arid Regions, 9(1): 78-88
- http://dx.doi.org/10.3724/SP.J.1226.2017.00078
Article History
- Received: August 30, 2016
- Accepted: November 12, 2016
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. Key Laboratory of Desert and Desertification, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu 730000, China;
4. Key Laboratory of Eco-hydrology and of Inland River Basin, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu 730000, China
Photosynthesis is not only the most important chemical reaction to transform inorganic matter into organic matter through fixing solar energy on the earth but also the crucial source for plant carbon or biomass accumulation for growth and development. Its productivity is tightly linked to the delicate balance between carboxylation and oxygenation. As a bifunctional enzyme catalyzing both the carboxylation of D-ribulose-1, 5-bisphosphate (RuBP) that initiates photosynthetic CO2 fixation and the oxygenation of RuBP that starts the nonessential photo-respiratory pathway (Nishimura et al., 2008), ribulose-1, 5-biphosphate carboxylase/oxygenase (Rubisco, EC4.1.1.39) plays a crucial role in the process of photosynthesis for most terrestrial plants. It consists of eight large subunit (LSU) and eight small subunits (SSU), compared to the little knowledge about the function of SSUs, LSUs have been proved to be the catalytic active site of the whole enzyme, which determines the catalytic efficiency; and the C-terminal of its amino acid was related to the fixation of CO2.
Rubisco has been proved to evolve toward high-efficiency utilization of CO2(Jordan and Ogren, 1981) for improving photosynthetic efficiency to adapt to environments (Galmes et al., 2005; Kubien et al., 2008). Previous studies showed that LSUs were encoded by the large single-copy region on the chloroplast genome, the rbcL gene (Strauss et al., 1988; Bausher et al., 2006). Although this gene was widely used as a molecular marker for the intergeneric or interfamilial level of phylogenetic analyses in angiosperms (Chase et al., 1993) with the characteristic of no evidence for adaptive evolution (Gould and Lewontin, 1979), as an important gene to encode the LSUs, rbcL was a likely target of natural selection to improve inefficient function of Rubisco, which brought the molecular evolution of the rbcL gene into question (Kapralov and Filatov, 2006). Recent researches showed that the evolution of the rbcL gene was indicated to be under positive selection in all principal lineages of land plants (Kapralov and Filatov, 2006). Furthermore, even under purifying selection in several species, such as Harveya purpurea, Striga gesnerioides, Orobanche fasciculata, and O. corymbosa (Wolfe and dePamphilis, 1997; Wolfe and dePamphilis, 1998; Leebens-Mack and DePamphilis, 2002). However, few studies focused on desert plants, which have faced extreme environment stresses such as the drought, UV, and high salt conditions for a long history.
Reaumuria soongarica (Tamaricaceae), a constructive xerophyte and halophyte shrub species, was widely distributed in all desert regions across the ACA (arid Central Asia) with an annual precipitation under 400 mm (Shi et al., 2013), including the Tengger Desert, Badain Jaran Desert, Gurbantunggut Desert, Gashun Gobi, Kumtag Desert, Qaidam Basin (on the northeastern Qinghai-Tibet Plateau, QTP), and Taklimakan Desert (which could be referred at www.eflora.org). Molecular phylogeographic study suggested that the evolutionary history of this Tertiary-relic species had been impacted by the uplift of the QTP and paleoclimate changes (Li et al., 2012; Yin et al., 2015). To be mentioned, as the typical C3 plants (Ma et al., 2007), a recent trancriptome of R. soongoricapresented a complete set of C4-function genes, which suggested that the regulation of Rubisco could be quite complicated for this plant to adapt to the extremely harsh environments. Furthermore, previous studies showed that the expression level and activase of the Rubisco were shifted in 142 land plants (Wang et al., 2011) under different environments, which also suggested a possible propensity to adapt to harsh habitats under different levels of salinity, water and temperature. Thus, it is reasonable to postulate that the rbcL subunit has undergone continual modification for better fitness in R. soongarica with shifts of paleogeography and paleoclimate.
In this study, to have a comprehensive acknowledge of whether the rbcL gene in R. soongarica experienced adaptive evolution, we first examined the sequence variations of the rbcL gene among 17 species of Tamaricaceae to seek the signal of adaptive evolution by applying phylogenetic analysis of maximum likelihood. Then, possible functional shifts were predicted by analysis of hydrophobicity and entropy on the locations under adaptive sites; the secondary and 3D-structure of Rubisco were also modelled to reveal higher levels of organization. Finally, to further clarify whether the expression level had shifted among different genotypes in R. soongorica in response to environment factors, the expression patterns of rbcL was analyzed under four treatments by real-time polymerase chain reaction (RT-PCR). Based on comprehensive analysis of the rbcL gene in R. soongorica, this study not only sheds light on the functional/structural features of Rubisco in R. soongorica but also provides useful information on directing genetic performance for enhancing the photosynthesis efficiency of desert plants to sustain fragile desert ecosystems, furthermore, to promote the ability to cope with desert aridification and global warming.
2 Material and methods 2.1 rbcL sequence acquirement and validation across biological databaseThe longest unigene with the annotation of ribulose-1, 5-biphosphate carboxylase/oxygenase was selected from the transcriptome of R. soongorica (Shi et al., 2013), and the peptide sequence was predicted on the GENSCAN Web Server at the Massachusetts Institute of Technology (MIT; http://genes.mit.edu/genscan.html). All the nucleotide sequences of rbcL from other species in Tamaricaceae were downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/). Finally, we obtained a total of 17 clean sequences (AY099899, AY099900, AY099902-AY099911, KC505169, KC505170, KC505172, Z 97650 and KM361003) in three genera, aligning with ClustalW implemented in Bioedit 7.2.2 (Hall, 1999). We also reconfirmed the ortholog gene of rbcL in R. soongorica by using the Basic Local Alignment Search Tool (BLAST) method on the Genebank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Rubisco-large superfamily conserved domain by using Pfam (http://pfam.xfam.org/).
2.2 Phylogenetic analysis and adaptive selection detectionTo confirm the phylogenetic relationships of rbcL in these 17 Tamaricaceae species, the maximum likelihood (ML) tree was reconstructed based on both the nucleotide and amino acid sequences analyses of rbcL using PhyML version 3.0 (Guindon and Gascuel, 2003) with 500 bootstraps under the best-fit model, respectively. The best-fit model for the nucleotide dataset was identified with the software of ModelGenerator, version 0.84 (Keane et al., 2004), based on Akaike information criterion (AIC) values (Akaike, 1974); and the TVM+I model and JTT+I model were finally chosen for nucleotide acid and amino acid sequences, respectively. Frankenia pulverulenta (HM850011) was used as the outgroup. Evolutionary changes and variations within and between species were judged with comparison of nonsynonymous-to-synonymous rates ratio (ω=dN/dS) by PAML package version 4.1 (Yang, 2007) with criteria as follows: when the ratio was equal (dN/dS=1), random drift of mutant alleles that are neutral at the molecular level was anticipated; when the ratio was lower than 1 (dN/dS < 1), purifying selection was expected; when the ratio was over 1 (dN/dS > 1), adaptive evolution or positive selection at the molecular level was indicated. The parameters of the M8 model using empirical Bayes approaches implemented in the CODEML program from PAML were used to calculate the posterior probabilities to identify amino acid sites potentially under positive selection.
2.3 Hydrophobicity and entropy analysisTo reveal the influence of mutation on biological diversity, and eventually contribute to the direction of biological diversity, the characteristics of the Shannon entropy of neighboring sites mutation were investigated with the program implemented in Bioedit 7.2.2 (Hall, 1999). Furthermore, to estimate the hydrophobicity force between polar AA side chains and the CO2 penetration into Rubisco's active site, hydrophobicity was also analyzed with the program implemented in Bioedit 7.2.2 (Hall, 1999).
2.4 Structural analysis of RubiscoAs an alternative approach for functional annotation of novel protein sequences, Phyre2 algorithm (http://www.sbg.bio.ic.ac.uk/phyre2/) was used to analyze the structure of Rubisco. The confidence score of Phyre2 was established with 55% as the minimum cut-off value, and the proteins with confidence scores equal to or higher than this cut-off value are shown. The pdb file from phyre2 was visually displayed by PyMOL software (Schrodinger, 2010), and positive selection sites were also marked with the PyMOL software editor.
2.5 Total RNA Isolation and cDNA Synthesis and Quantitative Real-time PCR (RT-PCR)To reveal the rbcL response to environmental factors, we germinated seeds of different geographic accession on Murashige-Skoog Medium (MS-medium) in the lab. After 14 days, the seedlings were exposed to drought, high-temperature, high-salt, and dark stresses. In detail, for drought stress, the seedlings were subjected to 15% PEG (polyethylene glycol) for 1 hour; for heat stress, the treatment was performed at 38 °C; for salt stress, plants were treated with 100 mmol/L NaCl for 3 hours; and for dark stress, plants were wrapped with foil paper for 2 hours. Total RNA of these materials was extracted using an E.Z.N.A. Plant RNA Kit (Omega Bio-tek, USA) with 2% PVBB added, and the remaining DNA was removed by RNase-free DNase (Omega Bio-tek, USA) according to the instruction manual. Total RNA concentration and purity was determined with ratio OD260/280 by NanoDrop1000 (Thermo Scientific), and all the samples passed quality control as the ratio of OD260/280 was between 1.9 to 2.2 and the ratio of OD260/230 < 2.0. RNA integrity was verified by 1.5% agarose gel electrophoresis with two clear bands of 28S/18S ribosomal RNA. For each sample, 1 μg of total RNA was reverse transcribed in a volume of 20 μL reaction with oligo dT primers by the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Germany). The cDNAs were diluted by 1:10 with nuclease-free water prior to the qPCR analyses.
RT-PCR primers for each unigene sequences fromde novo transcriptome ofTamarix hispida were designed by OligoPerfect Designer (http://tools.invitrogen.com/content.cfm?pageid=9716&icid=fr-oligo-6), with the length approximately 20 bp, optimal melting temperatures 60 °C, and optimal GC content 50%. Finally, two pairs of primers (rbcL and H2A) from Yan et al. (2014) with a product size between 100 to 300 bp were used in this study. PCR efficiency for each sample was calculated by LinRegPCR, with the mean PCR efficiency per amplicon between 1.8 and 2.0. A total of 20 μL reaction-system volume was used for amplification, consisting of 10 μL DyNAmo Flash SYBR Green qPCR Kit Master Mix (Thermo Scientific), 0.5 μL of forward and reverse primer, respectively, 0.2 μL of F-402 buffer, 2 μL cDNA synthesized from total RNA, and 6.8 μL double-distilled water. The PCR program contained an initial denaturation step of 5 min at 95 °C, followed by denaturation for 15 s at 95 °C, annealing for 30 s at 60 °C, and extension for 30 s at 72 °C for 40 cycles. The real-time PCR thermal cycler qTOWER 2.0/2.2 (Analytik Jena, Germany) was used to obtain relative expression levels of each sample. The dissociation curve was obtained by heating the amplicon from 60 °C to 95 °C for each primer to get good specificity and quality, efficiency, and the dissociation curve. The Cq value per sample and the fluorescence threshold was set as determined by LinRegPCR, and relative quantification was determined using the Delta Delta Ct method. The relative transcripts were normalized with reference genes from Yan et al. (2014).
3 Results 3.1 Gene identification and sequence analysisThe sequence of Unigene19142_A from the transcriptome of R. soongorica(Shi et al., 2013) was annotated as ribulose-1, 5-bisphosphate carboxylase/oxygenase large subunit. The predicted peptide on GENSCAN was 475 amino acids with 6 bp polyA at the position -289 to -284 and 5'-UTR at the position -2153 to -2114, which was similar to the previously published sequence of R. soongorica (AAM26908) with 100% identity, except for the decrease of 25 amino acids at the N-terminal and 28 amino acids at the C-terminal. Pfam results proved that the Rubisco-large superfamily was conserved even in different families (Figure 1).
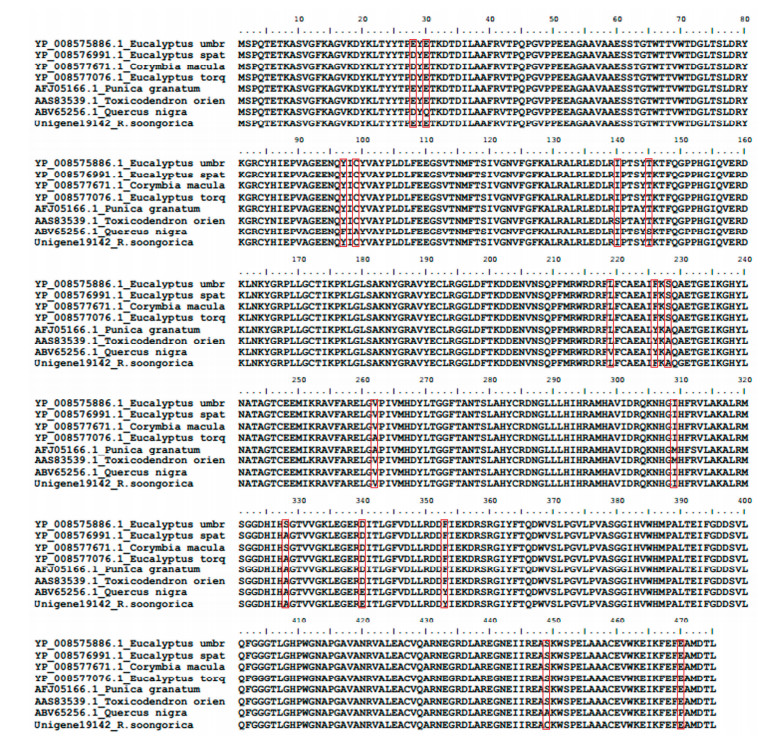
|
| Figure 1 Homologous sequences of Rubisco large subunit, with the red blocks representing the variation of amino acid of Rubisco large subunits in different species |
A total of 17 clean rbcL sequences with 1, 200 bp and their predicted protein sequences from Tamaricaceae, were used to reconstruct the nucleotide and protein phylogeny trees (Figure 2). As shown in Figure 2a, the tree is based on the variation of DNA sequences: the three genera, Reaumuria, Myricaria, and Tamarix, were independently separated from the main tree. In the positions of 336 and 595 of the DNA fragment, the mutations "T" and "G" were fixed in the Tamarix and Reaumuria genera, respectively, whereas, mutation "G" at position 660 was still in a steady-state fitness variation. However, the topology of the protein phylogeny tree was much different, as shown in Figure 2b; although the value of the bootstrap was low, Reaumuria was separated from the clade of Myricaria and Tamarix, and the species of these two genera were complicated to get a relatively clear lineage relationship. The discrepancy between the nucleotide and protein phylogeny trees suggested the level of differentiation in Tamaricaceae was higher in the transcript and translation process, which may contribute more to the divergence of Tamaricaceae species.
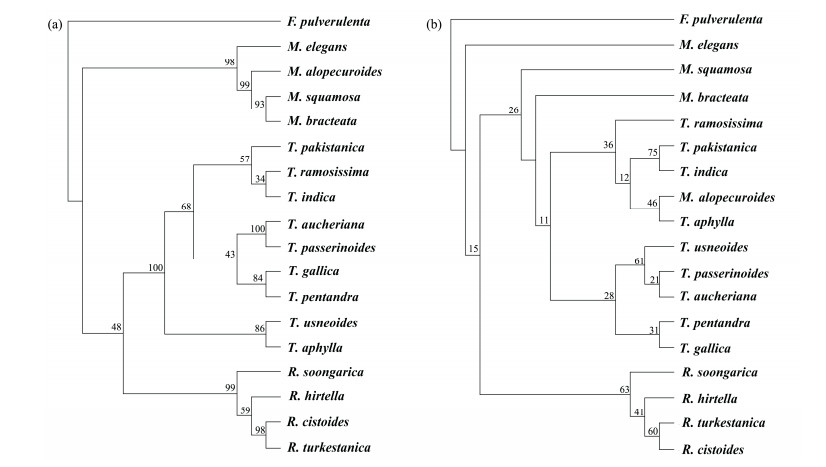
|
| Figure 2 Phylogenetic tree of Tamarixaceae. (a) phylogenetic tree reconstruct based on nucleotide acid sequences, (b) phylogenetic tree reconstruct based on amino acid sequences |
Results of the nonsynonymous/synonymous rate ratio ω showed that, with posterior probability over 95% by empirical Bayes analysis, three mutations at positions 112, 199, and 220 on rbcL were under positive selection (marked with an asterisk in Table 1). At position 112, the amino acid alanine was substituted by serine (Ala112→Ser112), which was largely harbored in the genera Tamarix and Reaumuria; at position 199, the amino acid threonine was replaced by alanine or glycine (Thr199→Ala199 or Thr199→ Gly199), which was largely harbored in genera Reaumuria and Myricaria; whereas, at position 220, the isoleucine was replaced by methionine (Ile220→ Met220), which was harbored in all the three genera (Table 2). As shown in Figure 1, all these substitutions were found in the terminal groups rather than the base group. According to the nucleotide sequences, the fixed "T" at position 336 was related to the amino acid mutation of Ala112→Ser112, the mutation "G" at 595 was related to the mutation of Thr199→Ala199 or Thr199→Gly199, while the complex mutation at 660 loci was clearly evolved from isoleucine to methionine under positive selection. Furthermore, the composite structure analysis showed that the sequence of R. soongorica harbored 11 substitutions, including eight non-selective positions and three selective positions (Figure 3a), and the 3D structure for Rubisco in R. soongorica showed that all of the three positive-selection positions (112, 199, and 220) were located in the alph-helix of the α/β barrel domain (red marker in Figure 3b).

|
| Figure 3 Predicted secondary and tertiary structures of the rbcL protein and substitution location. (a) the secondary structure of the rbcL protein and substitution location, with the nonselective positions marked with black boxes and the selective positions marked with purple boxes; (b) the tertiary structure of the rbcL protein and substitution location, with the alph-helix marked with red |
| Site | Pr (ω > 1) | Post mean | + - | SE for ω |
| 112A | 0.972* | 3.284 | + - | 1.062 |
| 114T | 0.851 | 2.938 | + - | 1.362 |
| 194L | 0.850 | 2.936 | + - | 1.362 |
| 195Y | 0.847 | 2.925 | + - | 1.368 |
| 199G | 0.972* | 3.283 | + - | 1.063 |
| 220I | 0.991** | 3.329 | + - | 1.003 |
| 289M | 0.756 | 2.631 | + - | 1.476 |
| 368V | 0.852 | 2.941 | + - | 1.360 |
| 384P | 0.849 | 2.933 | + - | 1.364 |
| * Significance lower than 0.05, ** significance lower than 0.01. The reference sequence was M. bracteata. | ||||
| Species | Positive-selection loci | ||
| 112 | 199 | 220 | |
| M. bracteata | A | T | I |
| M. squamosa | A | T | I |
| M. alopecuroides | A | T | M |
| M. elegans | A | A | I |
| R. cistoides | A | A | M |
| R. turkestanica | A | A | M |
| R. hirtella | A | A | I |
| R. soongarica | S | G | I |
| T. passerinoides | S | T | I |
| T. pakistanica | S | T | M |
| T. pentandra | S | T | I |
| T. indica | S | T | M |
| T. gallica | S | T | I |
| T. aphylla | S | T | M |
| T. usneoides | S | T | I |
| T. aucheriana | S | T | I |
| T. ramosissima | A | T | M |
From the Eisenberg scale, mean hydrophobicity analysis by Bioedit 7.2.2, the hydrophobicity decreased and polarity enhanced at the three selective loci (Figure 4a), suggesting that the hydrophobicity force between the polar AA side chains was shifted; and the conformation of the protein tends to be changed. Furthermore, the entropy was also increased, ranging from 0.6 to 1 (Figure 4b), also suggesting that those sites were unstable. The decreased hydrophobicity and increased entropy presumably facilitate CO2 penetration into Rubisco's active site, which suggests those selection loci were tightly impacted by a great change of CO2 concentration with the paleoclimate shift, to promote the evolution at the expense of a considerable energy cost to adapt to the environment.
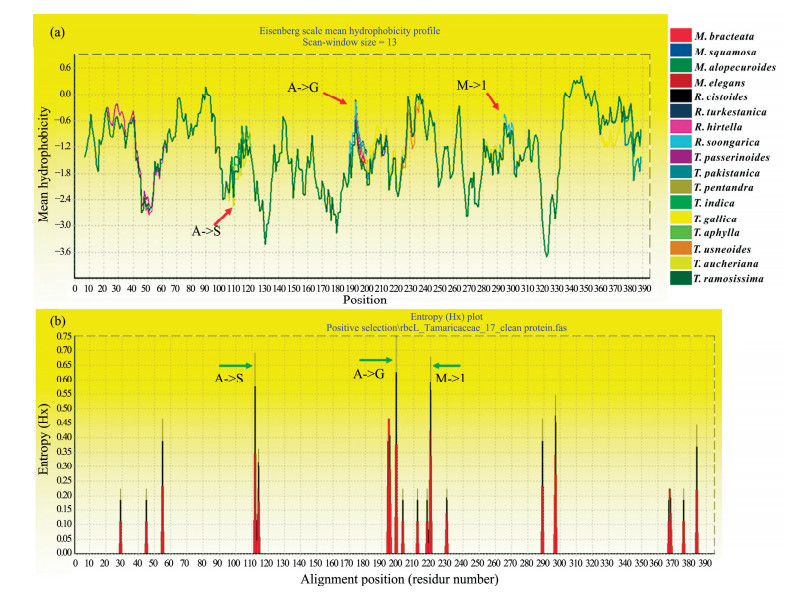
|
| Figure 4 Variation of hydrophobicity and entropy of rbcL sequence in R. soongorica. (a) variation of hydrophobicity on the rbcL sequence in R. soongorica, (b) entropy variation of positive-selection loci in R. soongorica |
Results of the RT-PCR showed that the average expression level of rbcL in the HG accessions was slightly upregulated under various stresses, while in the XGG accessions they were somewhat downregulated (Figure 5). Under the PEG treatment, the expression levels of rbcL were slightly increased and were higher in XGG accessions than HG ones. Under the heat treatment, the expression level of rbcL was slightly depressed or showed no change in HG accessions, whereas it was slightly increased in the accession of XGG. Under the dark treatment, the transcript level of rbcL remained unchanged except XGG030. Carbon assimilation is one of the major sites sensitive to drought, heat stress, and light in photosynthesis. The slight regulation of rbcL expression under PEG, heat, and dark treatments revealed that the maintenance mechanism of CO2-assimilation capacity was mainly protected by Rubisco activity or degradation of Rubisco. Different from the other three accessions, the HG020 elevated to a high expression level under the salt treatment. This finding could be attributed to both the genetic variation in the local environment and the regulation of rbcL expression.
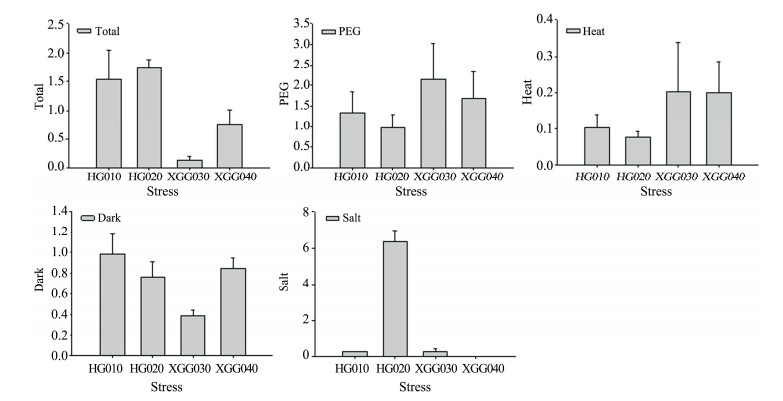
|
| Figure 5 rbcL gene expression under various stresses |
Although, in the field of phylogenetic analyses, the rbcL gene has been considered to be conserved (Chase et al., 1993), issues of rbcL evolution under natural selection had been proposed recently within all principal lineages of land plants. Most of the Rubisco residues appeared to be under purifying selection (Wolfe and dePamphilis, 1997; Wolfe and dePamphilis, 1998; Leebens-Mack and DePamphilis, 2002), while only a small fraction have been suggested to be under positive selection in particular taxonomic groups (Kapralov and Filatov, 2006; Christin et al., 2008; Iida et al., 2009; Miwa et al., 2009; Sen et al., 2011; Wang et al., 2011; Young et al., 2012). These phenomena indicated that the evolution of rbcL, which was induced by the constant fine-tuning of Rubisco to adapt to different environments, was always occurring (Galmés et al., 2005).
In this study, three nonsynonymous mutations were suggested to be under positive selection in the family Tamaricaceae (Table 1). They were harbored in the terminal groups rather than the base group (Figure 1); and the fixed mutation of "T" at position 336 contributed to the Ala112→Ser112; the mutation "G" at position 595 contributed to Thr199→Ala199 or Thr199→ Gly199; and the flexible mutation at position 220 was evolved from isoleucine to methionine under positive selection. On the level of nucleotide acid, the mutations "T" and "G" were fixed in Tamarix and Reaumuria genera, respectively, whereas, mutation "G" at loci 660 was still in a steady-state fitness variation, which contributed to the independent clade for the three genera, Reaumuria, Myricaria, and Tamarix. On the level of amino acid, Ala112→Ser112was harbored majorly in genera Tamarix and Reaumuria; Thr199→ Ala199 or Thr199→Gly199 was harbored majorly in genera Reaumuria and Myricaria, whereas, Ile220→ Met220 was harbored in all three genera. In considering their habitat environments, we suggest that these replacements may account for the ecological differences; and Ala112→Ser112seemed to be more responsible for the drought-and salinity-selection pressure, as water is demanded for increasing survival and the salt-secreting ability was reduced from Reaumuria to Tamarix toMyricaria. The spatial analysis showed that all the three positive-selection loci (112, 199, and 220) were located in the alph-helix (red marker in Figure 3b), and previous studies showed that the positively selected sites preferably located in the helices (Wang et al., 2011). Furthermore, we also found that these substitutions were introduced to the decrease of hydrophobicities and the increase of the entropy (Figure 4), supporting the contention that these selective substitutions could induce the instability of the Rubisco protein structure and contribute to the diversity of species. According to Yin et al. (2015), the intrafamily divergence of Tamaraceae was dated to approximately 66.36 Ma, and the two genera Tamarix and Myricaria could have diverged at approximately 43.26 Ma, when a higher concentration of CO2 was in the atmosphere. The high concentration of CO2 induced the faster evolution of Rubisco to more efficient utilization of CO2 (Jordan and Ogren, 1981; Savir et al., 2010). As an example, R. soongarica had been regarded as the Tertiary-relic species (Hou, 1987), which represented the background of a great shift in global CO2 concentration (Petit et al., 1999; Zachos et al., 2001) around Rubisco. At that time, C4 plants arose from C3 plants via the gradual addition of constituents and their ability to thrive across a diversity of habitats (Christin and Osborne, 2013). The variation of δ13C value for R. soongorica ranged from -22.8‰ to -29.9‰, and the lowest δ13C appeared to favor salinity. What's more, the values increase when the salinity is lower or higher than the optimum level; and the variation of δ13C may be attributed to all of the genes encoding key enzymes in the C4carbon-fixation pathway, detected in the transcriptomic dataset by Shi et al. (2013); thus, we would like to infer that there may exist the intermediate of C3-C4 photosynthesis in R. soongorica to adapt to different environments, as found in other species (Ueno, 1998; Brautigam et al., 2011; Williams et al., 2012).
Environmental factors such as salinity, temperature, and drought could induce the limitation of CO2 fixation and further result in the decrease of photosynthetic capacity in plants (Barhoumi et al., 2007; Hozain et al., 2010; Carmo-Silva and Salvucci, 2012; Du et al., 2015; Guo et al., 2015), which were suggested to be a driving force for the evolution of Rubiscio (Jordan and Ogren, 1981; Delgado et al., 1995; Galmes et al., 2015). As found in other plants such as the soybean (Carmo-Silva et al., 2010) and tomato (Wang et al., 2015), the transcriptional levels of rbcL in R. soongorica, which could reflect the maintenance of CO2-assimilation capacity for plants, was slightly regulated by dark, heat, and PEG treatments (Figure 5). However, the response to the salt treatment was complex, as shown in Figure 5, with the expression level of rbcL highly upregulated in HG020 accession, while no change or slight deregulation in the other three accessions. In fact, the expression level of rbcL is not only associated with the concentration of Rubisco (Randle and Wolfe, 2005; Mehrotra et al., 2011) but also with the activation state of the enzyme (Galmes et al., 2013) and the synthesis and degradation of Rubisco or the regulation mechanism (Mori et al., 2002; Bowman et al., 2013; Wang et al., 2015). Until now, the precise modulation of Rubisco activity under varying environmental conditions was unclear, such as different responses to drought stress were found in different species (Sharkey and Seemann, 1989; Parry et al., 2002; Galmes et al., 2013; Zhou et al., 2015).
Numerous phylogeographic studies have suggested that natural selection would drive adaptive diversification of genes (Liu and Zhu, 2010; Zhao et al., 2014; Huang et al., 2015; Sun et al., 2015) and even result in intraspecific variation (Fu et al., 2013; Johnson et al., 2015; Yin et al., 2015). A great genetic variability in salt tolerance among genotypes of species had been shown (Lowry et al., 2009; Bchini et al., 2010), and salinity tolerance was positively related to the genetic diversity of species (Zhang and Chen, 2010). In this study, we found that the expression level of the Rubisco and its activase were shifted in response to variation of CO2 concentration in the history; furthermore, the heterogeneous desert environments may also contribute to the functional divergence of Rubisco. To have a deeper understanding of the adaptive evolution of rbcL, further experiments with wider sampling are still needed. At present, this study presented an example of the way that Tamaricaceae plants orchestrate the C3 and C4 pathways to adapt to environmental changes, and it further sheds light on the evolution scenarios from the C3-like pathway to the C4 pathway.
Acknowledgments:This work was supported by the National Natural Science Foundation of China (NSFC, Grant Nos. 31370395 and 31500266) and the "One Hundred Talents" project of the Chinese Academy of Sciences (Grant No. 29Y127E71).
| Akaike H, 1974. A new look at the statistical model identification. Automatic Control, IEEE Transactions, 19: 716–723. DOI: 10.1109/TAC.1974.1100705 |
| Barhoumi Z, Djebali W, Chaibi W, et al, 2007. Salt impact on photosynthesis and leaf ultrastructure of Aeluropus littoralis. Journal of Plant Research, 120(4): 529–537. DOI: 10.1007/s10265-007-0094-z |
| Bausher MG, Singh ND, Lee SB, et al, 2006. The complete chlo-roplast genome sequence of Citrus sinensis (L. ) Osbeck var Ridge Pineapple:organization and phylogenetic relationships to other angiosperms. BMC Plant Biology, 6: 21. |
| Bchini H, Ben Naceur M, Sayar R, et al, 2010. Genotypic differ-ences in root and shoot growth of barley (Hordeum vulgare L. ) grown under different salinity levels. Hereditas, 147(3): 114–122. |
| Bowman SM, Patel M, Yerramsetty P, et al, 2013. A novel RNA binding protein affects rbcL gene expression and is specific to bundle sheath chloroplasts in C4 plants. BMC Plant Biology, 13: 138. DOI: 10.1186/1471-2229-13-138 |
| Brautigam A, Kajala K, Wullenweber J, et al, 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology, 155(1): 142–156. DOI: 10.1104/pp.110.159442 |
| Carmo-Silva AE, Keys AJ, Andralojc PJ, et al, 2010. Rubisco activities, properties, and regulation in three different C4 grasses under drought. Journal of Experimental Botany, 61(9): 2355–2366. DOI: 10.1093/jxb/erq071 |
| Carmo-Silva AE, Salvucci ME, 2012. The temperature response of CO2 assimilation, photochemical activities and Rubisco acti-vation in Camelina sativa, a potential bioenergy crop with lim-ited capacity for acclimation to heat stress. Planta, 236(5): 1433–1445. DOI: 10.1007/s00425-012-1691-1 |
| Chase MW, Soltis DE, Olmstead RG, et al, 1993. Phylogenetics of seed plants:an analysis of nucleotide sequences from the plastid gene rbcL. Annals of the Missouri Botanical Garden, 80(3): 528–580. DOI: 10.2307/2399846 |
| Christin PA, Osborne CP, 2013. The recurrent assembly of C4 photosynthesis, an evolutionary tale. Photosynthesis Research, 117(1-3): 163–175. DOI: 10.1007/s11120-013-9852-z |
| Christin PA, Salamin N, Muasya AM, et al, 2008. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Molecular Biology and Evolution, 25(11): 2361–2368. DOI: 10.1093/molbev/msn178 |
| Delgado E, Medrano H, Keys AJ, et al, 1995. Species variation in Rubisco specificity factor. Journal of Experimental Botany, 46(292): 1775–1777. |
| Du QJ, Dai KR, Li JM, et al, 2015. Effects of sub-low temperature and drought stress on characteristics of photosynthetic and fluorescence kinetics in tomato leaves. The Journal of Applied Ecology, 26(6): 1687–1694. |
| Fu H, Yuan G, Zhong J, et al, 2013. Environmental and ontogenetic effects on intraspecific trait variation of a macrophyte species across five ecological scales. PLoS ONE, 8(4): e62794. DOI: 10.1371/journal.pone.0062794 |
| Galmes J, Aranjuelo I, Medrano H, et al, 2013. Variation in Ru-bisco content and activity under variable climatic factors. Photosynthesis Research, 117(1-3): 73–90. DOI: 10.1007/s11120-013-9861-y |
| Galmes J, Flexas J, Keys A, et al, 2005. Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant, Cell and Environment, 28: 9. |
| Galmés J, Flexas J, Keys AJ, et al, 2005. Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant, Cell & Environment, 28: 571–579. |
| Galmes J, Kapralov MV, Copolovici LO, et al, 2015. Temperature responses of the Rubisco maximum carboxylase activity across domains of life:phylogenetic signals, trade-offs, and im-portance for carbon gain. Photosynthesis Research, 123(2): 183–201. DOI: 10.1007/s11120-014-0067-8 |
| Gould SJ, Lewontin RC, 1979. The spandrels of San Marco and the Panglossian paradigm:a critique of the adaptationist pro-gramme. Proceedings of the Royal Society of London B:Bio-logical Sciences, 205: 581–598. DOI: 10.1098/rspb.1979.0086 |
| Guindon S, Gascuel O, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52: 696–704. DOI: 10.1080/10635150390235520 |
| Guo SJ, Yang KM, Huo J, et al, 2015. Influence of drought on leaf photosynthetic capacity and root growth of soybeans at grain filling stage. The Journal of Applied Ecology, 26(5): 1419–1425. |
| Hall TA, 1999. BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. |
| Hou X, 1987. Geographical distribution of vegetation in arid desert area of Chinese temperate zones. Botany Bulletin,: 37–66. |
| Hozain MI, Salvucci ME, Fokar M, et al, 2010. The differential response of photosynthesis to high temperature for a boreal and temperate Populus species relates to differences in Rubisco activation and Rubisco activase properties. Tree Physiology, 30(1): 32–44. DOI: 10.1093/treephys/tpp091 |
| Huang Y, Wang X, Ge S, et al, 2015. Divergence and adaptive evolution of the gibberellin oxidase genes in plants. BMC Evolutionary Biology, 15(1): 1–15. DOI: 10.1186/s12862-014-0274-0 |
| Iida S, Miyagi A, Aoki S, et al, 2009. Molecular adaptation of rbcL in the heterophyllous aquatic plant Potamogeton. PLoS ONE, 4(2): e4633. DOI: 10.1371/journal.pone.0004633 |
| Johnson LC, Olsen JT, Tetreault H, et al, 2015. Intraspecific variation of a dominant grass and local adaptation in reciprocal garden communities along a US Great Plains' precipitation gradient:implications for grassland restoration with climate change. Evolutionary Applications, 8(7): 705–723. DOI: 10.1111/eva.12281 |
| Jordan DB, Ogren WL, 1981. Species variation in the specificity of ribulose biphosphate carboxylase/oxygenase. Nature, 291(5815): 513–515. DOI: 10.1038/291513a0 |
| Kapralov MV, Filatov DA, 2006. Molecular adaptation during adaptive radiation in the Hawaiian endemic genus Schiedea. PLoS ONE, 1: e8. DOI: 10.1371/journal.pone.0000008 |
| Kapralov MV, Kubien DS, Andersson I, et al, 2011. Changes in Rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Molecular Biology and Evolution, 28(4): 1491–1503. DOI: 10.1093/molbev/msq335 |
| Keane T, Naughton T, McInerney J, 2004. ModelGenerator:amino acid and nucleotide substitution model selection. National University of Ireland, Maynooth, Ireland, 34. |
| Kubien DS, Whitney SM, Moore PV, et al, 2008. The biochemistry of Rubisco in Flaveria. Journal of Experimental Botany, 59(7): 1767–1777. |
| Leebens-Mack J, DePamphilis C, 2002. Power analysis of tests for loss of selective constraint in cave crayfish and nonphotosyn-thetic plant lineages. Molecular Biology and Evolution, 19(8): 1292–1302. DOI: 10.1093/oxfordjournals.molbev.a004190 |
| Li ZH, Chen J, Zhao GF, et al, 2012. Response of a desert shrub to past geological and climatic change:A phylogeographic study of Reaumuria soongarica (Tamaricaceae) in western China. Journal of Systematics and Evolution, 50(4): 351–361. DOI: 10.1111/j.1759-6831.2012.00201.x |
| Liu Q, Zhu Z, 2010. Functional divergence of the NIP III subgroup proteins involved altered selective constraints and positive se-lection. BMC Plant Biology, 10(1): 1–12. DOI: 10.1186/1471-2229-10-1 |
| Lowry DB, Hall MC, Salt DE, et al, 2009. Genetic and physio-logical basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. The New Phytologist, 183(3): 776–788. DOI: 10.1111/nph.2009.183.issue-3 |
| Ma JY, Chen K, Xia DS, et al, 2007. Variation in foliar stable carbon isotope among populations of a desert plant, Reaumuria soongorica (Pall. ) Maxim. in different environments. Journal of Arid Environments, 69(3): 365–374. |
| Mehrotra S, Trivedi PK, Sethuraman A, et al, 2011. The rbcL gene of Populus deltoides has multiple transcripts and is re-dox-regulated in vitro. Journal of Plant Physiology, 168(5): 466–473. DOI: 10.1016/j.jplph.2010.08.001 |
| Miwa H, Odrzykoski IJ, Matsui A, et al, 2009. Adaptive evolution of rbcL in Conocephalum (Hepaticae, bryophytes). Gene, 441(1-2): 169–175. DOI: 10.1016/j.gene.2008.11.020 |
| Mori S, Castoreno A, Lammers PJ, 2002. Transcript levels of rbcR1, ntcA, and rbcL/S genes in cyanobacterium Anabaena sp. PCC 7120 are downregulated in response to cold and osmotic stress. FEMS Microbiology Letters, 213(2): 167–173. |
| Nishimura K, Ogawa T, Ashida H, et al, 2008. Molecular mecha-nisms of Rubisco biosynthesis in higher plants. Plant Biotech-nology, 25(3): 285–290. DOI: 10.5511/plantbiotechnology.25.285 |
| Parry MA, Andralojc PJ, Khan S, et al, 2002. Rubisco activity:effects of drought stress. Annals of Botany, 89(Spec.): 833–839. |
| Petit JR, Jouzel J, Raynaud D, et al, 1999. Climate and atmospheric history of the past 420, 000 years from the Vostok ice core, Antarctica. Nature, 399(6735): 429–436. DOI: 10.1038/20859 |
| Randle CP, Wolfe AD, 2005. The evolution and expression of RBCL in holoparasitic sister-genera Harveya and Hyobanche (Orobanchaceae). American Journal of Botany, 92(9): 1575–1585. DOI: 10.3732/ajb.92.9.1575 |
| Savir Y, Noor E, Milo R, et al, 2010. Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proceedings of the National Academy of Sciences of the United States of America, 107(8): 3475–3480. DOI: 10.1073/pnas.0911663107 |
| Schrodinger LLC, 2010. The AxPyMOL Molecular Graphics Plugin for Microsoft PowerPoint, Version 1.0. |
| Sen L, Fares MA, Liang B, et al, 2011. Molecular evolution of rbcL in three gymnosperm families:identifying adaptive and coevolutionary patterns. Biology Direct, 6: 29. DOI: 10.1186/1745-6150-6-29 |
| Sharkey TD, Seemann JR, 1989. Mild water stress effects on carbon-reduction-cycle intermediates, ribulose bisphosphate carboxylase activity, and spatial homogeneity of photosynthesis in intact leaves. Plant physiology, 89(4): 1060–1065. DOI: 10.1104/pp.89.4.1060 |
| Shi Y, Yan X, Zhao P, et al, 2013. Transcriptomic analysis of a tertiary relict plant, extreme xerophyte Reaumuria soongorica to identify genes related to drought adaptation. PLoS ONE, 8(5): e63993. DOI: 10.1371/journal.pone.0063993 |
| Strauss SH, Palmer JD, Howe GT, et al, 1988. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proceedings of the National Academy of Sciences, 85(11): 3898–3902. DOI: 10.1073/pnas.85.11.3898 |
| Sun H, Guo K, Feng S, et al, 2015. Positive selection drives adaptive diversification of the 4-coumarate:CoA ligase (4CL) gene in angiosperms. Ecology and Evolution, 5(16): 3413–3420. DOI: 10.1002/ece3.2015.5.issue-16 |
| Tcherkez GG, Farquhar GD, Andrews TJ, 2006. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proceedings of the National Academy of Sciences of the United States of America, 103(19): 7246–7251. DOI: 10.1073/pnas.0600605103 |
| Ueno O, 1998. Induction of kranz anatomy and C4-like biochemical characteristics in a submerged amphibious plant by abscisic acid. The Plant Cell, 10(4): 571–584. |
| Wang G, Kong F, Zhang S, et al, 2015. A tomato chloro-plast-targeted DnaJ protein protects Rubisco activity under heat stress. Journal of Experimental Botany, 66(11): 3027–3040. DOI: 10.1093/jxb/erv102 |
| Wang M, Kapralov M, Anisimova M, 2011. Coevolution of amino acid residues in the key photosynthetic enzyme Rubisco. BMC Evolutionary Biology, 11(1): 266. DOI: 10.1186/1471-2148-11-266 |
| Williams BP, Aubry S, Hibberd JM, 2012. Molecular evolution of genes recruited into C4 photosynthesis. Trends in Plant Science, 17(4): 213–220. DOI: 10.1016/j.tplants.2012.01.008 |
| Wolfe AD, dePamphilis CW, 1997. Alternate paths of evolution for the photosynthetic gene rbcL in four nonphotosynthetic species of Orobanche. Plant Molecular Biology, 33(6): 965–977. DOI: 10.1023/A:1005739223993 |
| Wolfe AD, dePamphilis CW, 1998. The effect of relaxed functional constraints on the photosynthetic gene rbcL in photosynthetic and nonphotosynthetic parasitic plants. Molecular Biology and Evolution, 15(10): 1243–1258. DOI: 10.1093/oxfordjournals.molbev.a025853 |
| Yan X, Dong X, Zhang W, et al, 2014. Reference gene selection for quantitative real-time PCR normalization in Reaumuria soon-gorica. PLoS ONE, 9(8): e104124. DOI: 10.1371/journal.pone.0104124 |
| Yang Z, 2007. PAML 4:phylogenetic analysis by maximum like-lihood. Molecular Biology and Evolution, 24(8): 1586–1591. DOI: 10.1093/molbev/msm088 |
| Yin H, Yan X, Shi Y, et al, 2015. The role of East Asian monsoon system in shaping population divergence and dynamics of a constructive desert shrub Reaumuria soongarica. Scientific Reports, 5: 15823. DOI: 10.1038/srep15823 |
| Young JN, Rickaby RE, Kapralov MV, et al, 2012. Adaptive signals in algal Rubisco reveal a history of ancient atmospheric carbon dioxide. Philosophical transactions of the Royal Society of London (Series B:Biological Sciences), 367(1588): 483–492. DOI: 10.1098/rstb.2011.0145 |
| Zachos J, Pagani M, Sloan L, et al, 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science, 292(5517): 686–693. DOI: 10.1126/science.1059412 |
| Zhang JY, Chen JX C, 2010. Salinity tolerance and genetic diversity of the dinoflagllate Oxyrrhis marina. Journal of Ocean Uni-versity of China, 9(1): 87–93. DOI: 10.1007/s11802-010-0087-8 |
| Zhao Z, Liu H, Luo Y, et al, 2014. Molecular evolution and functional divergence of tubulin superfamily in the fungal tree of life. Scientific Reports, 4: 6746. DOI: 10.1038/srep06746 |
| Zhou SX, Medlyn BE, Prentice IC, 2015. Long-term water stress leads to acclimation of drought sensitivity of photosynthetic capacity in xeric but not riparian Eucalyptus species. Annals of Botany, 117(1): 133–144. |
 2017, 9
2017, 9


